bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1658_orf1
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
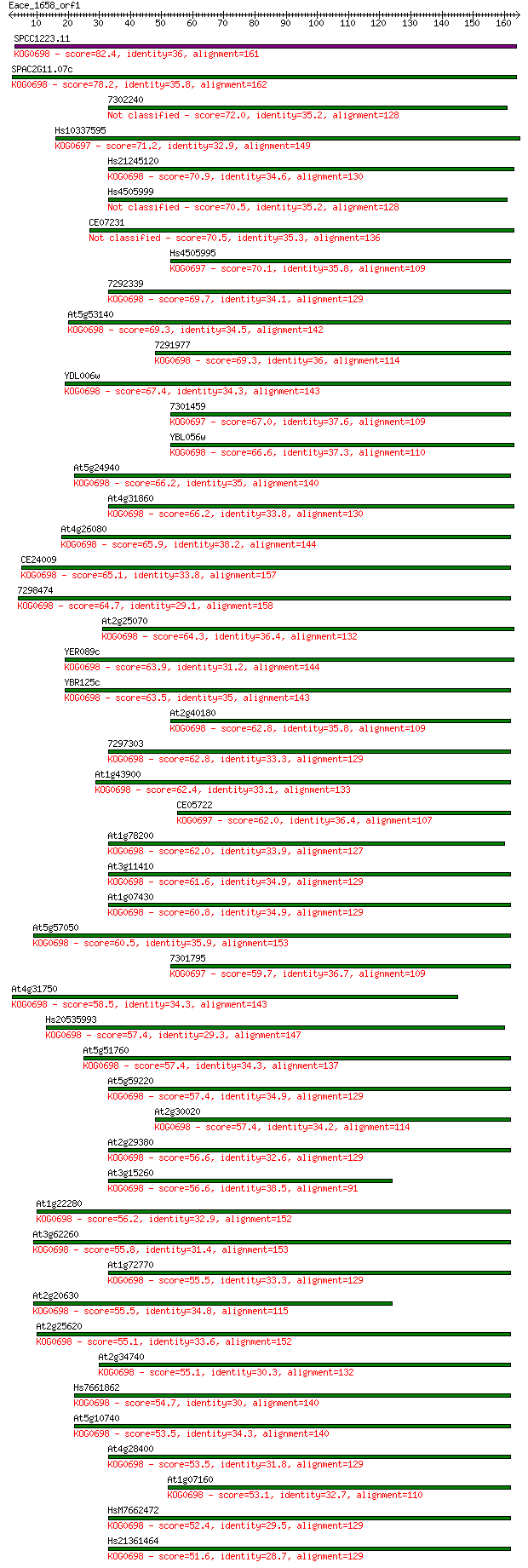
SPCC1223.11 82.4 4e-16
SPAC2G11.07c 78.2 7e-15
7302240 72.0 5e-13
Hs10337595 71.2 9e-13
Hs21245120 70.9 1e-12
Hs4505999 70.5 1e-12
CE07231 70.5 2e-12
Hs4505995 70.1 2e-12
7292339 69.7 3e-12
At5g53140 69.3 3e-12
7291977 69.3 3e-12
YDL006w 67.4 1e-11
7301459 67.0 2e-11
YBL056w 66.6 2e-11
At5g24940 66.2 2e-11
At4g31860 66.2 3e-11
At4g26080 65.9 4e-11
CE24009 65.1 6e-11
7298474 64.7 8e-11
At2g25070 64.3 1e-10
YER089c 63.9 1e-10
YBR125c 63.5 2e-10
At2g40180 62.8 3e-10
7297303 62.8 3e-10
At1g43900 62.4 4e-10
CE05722 62.0 5e-10
At1g78200 62.0 5e-10
At3g11410 61.6 7e-10
At1g07430 60.8 1e-09
At5g57050 60.5 1e-09
7301795 59.7 2e-09
At4g31750 58.5 5e-09
Hs20535993 57.4 1e-08
At5g51760 57.4 1e-08
At5g59220 57.4 1e-08
At2g30020 57.4 1e-08
At2g29380 56.6 2e-08
At3g15260 56.6 2e-08
At1g22280 56.2 3e-08
At3g62260 55.8 3e-08
At1g72770 55.5 4e-08
At2g20630 55.5 5e-08
At2g25620 55.1 5e-08
At2g34740 55.1 6e-08
Hs7661862 54.7 9e-08
At5g10740 53.5 2e-07
At4g28400 53.5 2e-07
At1g07160 53.1 2e-07
HsM7662472 52.4 4e-07
Hs21361464 51.6 6e-07
> SPCC1223.11
Length=370
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 58/167 (34%), Positives = 82/167 (49%), Gaps = 18/167 (10%)
Query 3 MPDFSGNSFAEA----SFALDAALRSDPEIPIGP-GSTGIIGILLAEDPAVPEFTLHVAN 57
P F ++ EA A D AL D ++ P G T ++ V ++ AN
Sbjct 85 QPSFWKGNYDEALKSGFLAADNALMQDRDMQEDPSGCTATTALI------VDHQVIYCAN 138
Query 58 IGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSF 117
GDSR VL GT P+S D KPN+ VE AR+ AGG + R++G LALSR+
Sbjct 139 AGDSRT-VLGRKGTAEPLSFDHKPNNDVEKARITAAGGFI-----DFGRVNGSLALSRAI 192
Query 118 GDFRMKGRTDILPSQQKVVALPHARTIKVKAGD-ILVFACDGVFEAQ 163
GDF K + + P +Q V A P + D L+ ACDG+++ +
Sbjct 193 GDFEYKKDSSLPPEKQIVTAFPDVVIHNIDPDDEFLILACDGIWDCK 239
> SPAC2G11.07c
Length=414
Score = 78.2 bits (191), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 58/168 (34%), Positives = 84/168 (50%), Gaps = 20/168 (11%)
Query 2 RMPDFSGNSFAEA---SF-ALDAALRSDPEIPIGP-GSTGIIGILLAEDPAVPEFTLHVA 56
+ PDF F A SF D A+ D + P G T + + + L+ A
Sbjct 83 KNPDFQKGDFVNALKSSFLNADKAILDDDQFHTDPSGCTATVVLRVGN-------KLYCA 135
Query 57 NIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRS 116
N GDSR VL + G P+S D KP++ E AR+ AGG V R++G LALSR+
Sbjct 136 NAGDSRT-VLGSKGIAKPLSADHKPSNEAEKARICAAGGFV-----DFGRVNGNLALSRA 189
Query 117 FGDFRMKGRTDILPSQQKVVALPHARTIKVKAGD-ILVFACDGVFEAQ 163
GDF K +++ P +Q V ALP ++ D +V ACDG+++ +
Sbjct 190 IGDFEFK-NSNLEPEKQIVTALPDVVVHEITDDDEFVVLACDGIWDCK 236
> 7302240
Length=662
Score = 72.0 bits (175), Expect = 5e-13, Method: Composition-based stats.
Identities = 45/129 (34%), Positives = 66/129 (51%), Gaps = 13/129 (10%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
G T ++ +L D L+VAN GDSR V+ G MS+D KP D E +R+ +
Sbjct 393 GCTAVVCLLQGRD-------LYVANAGDSRC-VISRSGQAIEMSIDHKPEDDEEASRIIK 444
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAGD-I 151
AGG V R++G L LSR+ GD K + +Q + ALP + + + D
Sbjct 445 AGGRVTLDG----RVNGGLNLSRALGDHAYKTNVTLPAEEQMISALPDIKKLIITPEDEF 500
Query 152 LVFACDGVF 160
+V ACDG++
Sbjct 501 MVLACDGIW 509
> Hs10337595
Length=382
Score = 71.2 bits (173), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 49/153 (32%), Positives = 77/153 (50%), Gaps = 16/153 (10%)
Query 16 FALDAALRSDPEIPIGPGSTG--IIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFT 73
+D +R E G +G +G+L++ P+ T + N GDSR L+
Sbjct 106 LEIDEHMRVMSEKKHGADRSGSTAVGVLIS-----PQHTYFI-NCGDSRGLLCRNRKVHF 159
Query 74 PMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQ 133
+ D KP++P+E R+Q AGGSVM + R++G LA+SR+ GDF K P++Q
Sbjct 160 -FTQDHKPSNPLEKERIQNAGGSVM-----IQRVNGSLAVSRALGDFDYKCVHGKGPTEQ 213
Query 134 KVVALPHARTIKVKAGD--ILVFACDGVFEAQG 164
V P I+ D ++ ACDG+++ G
Sbjct 214 LVSPEPEVHDIERSEEDDQFIILACDGIWDVMG 246
> Hs21245120
Length=181
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 70/132 (53%), Gaps = 18/132 (13%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
G+T +I +L +D L VAN+GDSR ++ DG P+S D KP E R++R
Sbjct 13 GTTCLIALLSDKD-------LTVANVGDSRGVLCDKDGNAIPLSHDHKPYQLKERKRIKR 65
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKA--GD 150
AGG + + G WR+ G LA+SRS GD+ +K ++P P T + +
Sbjct 66 AGGFI--SFNGSWRVQGILAMSRSLGDYPLKNLNVVIPD-------PDILTFDLDKLQPE 116
Query 151 ILVFACDGVFEA 162
++ A DG+++A
Sbjct 117 FMILASDGLWDA 128
> Hs4505999
Length=546
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 45/129 (34%), Positives = 69/129 (53%), Gaps = 13/129 (10%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
G+T ++ ++ + L VAN GDSR +V G MS D KP D VE+AR++
Sbjct 328 GTTAVVALIRGKQ-------LIVANAGDSRCVVSEA-GKALDMSYDHKPEDEVELARIKN 379
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG-DI 151
AGG V R++G L LSR+ GD K ++ P +Q + ALP + + + +
Sbjct 380 AGGKVTMDG----RVNGGLNLSRAIGDHFYKRNKNLPPEEQMISALPDIKVLTLTDDHEF 435
Query 152 LVFACDGVF 160
+V ACDG++
Sbjct 436 MVIACDGIW 444
> CE07231
Length=491
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 71/140 (50%), Gaps = 19/140 (13%)
Query 27 EIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVE 86
E+P T L+ +D + VAN GDSR VL +G +SVD KP D VE
Sbjct 309 EVPGEDSGTTACVCLVGKDKVI------VANAGDSRA-VLCRNGKAVDLSVDHKPEDEVE 361
Query 87 MARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKV 146
R+ AGG + R++G L LSR+FGD K ++ +Q + ALP +K+
Sbjct 362 TNRIHAAGGQIEDG-----RVNGGLNLSRAFGDHAYKKNQELGLKEQMITALPD---VKI 413
Query 147 KA----GDILVFACDGVFEA 162
+A + +V ACDG++ +
Sbjct 414 EALTPEDEFIVVACDGIWNS 433
> Hs4505995
Length=479
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 62/110 (56%), Gaps = 7/110 (6%)
Query 53 LHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLA 112
++ N GDSR VL+ +G + D KP +P E R+Q AGGSVM + R++G LA
Sbjct 144 IYFINCGDSRA-VLYRNGQVCFSTQDHKPCNPREKERIQNAGGSVM-----IQRVNGSLA 197
Query 113 LSRSFGDFRMKGRTDILPSQQKVVALPHARTI-KVKAGDILVFACDGVFE 161
+SR+ GD+ K P++Q V P I + + + ++ ACDG+++
Sbjct 198 VSRALGDYDYKCVDGKGPTEQLVSPEPEVYEILRAEEDEFIILACDGIWD 247
> 7292339
Length=371
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 44/130 (33%), Positives = 67/130 (51%), Gaps = 15/130 (11%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST ++ + V + L+ AN GDSR + +G +S+D KPN+ E R+ +
Sbjct 115 GSTAVVVL-------VKDNKLYCANAGDSRAIAC-VNGQLEVLSLDHKPNNEAESKRIIQ 166
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKV-KAGDI 151
GG V R++G LALSR+ GD+ K + P Q V A P T K+ +
Sbjct 167 GGGWVEFN-----RVNGNLALSRALGDYVFK-HENKKPEDQIVTAFPDVETRKIMDDWEF 220
Query 152 LVFACDGVFE 161
+V ACDG+++
Sbjct 221 IVLACDGIWD 230
> At5g53140
Length=307
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 49/143 (34%), Positives = 74/143 (51%), Gaps = 20/143 (13%)
Query 20 AALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQ 79
A L S+ + GST +L+ L+VAN+GDSR +V G +S D
Sbjct 67 AFLESEKDTYRDDGSTASAAVLVGNH-------LYVANVGDSRTIVSKA-GKAIALSDDH 118
Query 80 KPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALP 139
KPN E R++ AGG +M G WR+ G LA+SR+FG+ +K Q VVA P
Sbjct 119 KPNRSDERKRIESAGGVIMWA--GTWRVGGVLAMSRAFGNRMLK---------QFVVAEP 167
Query 140 HARTIKV-KAGDILVFACDGVFE 161
+ +++ ++LV A DG+++
Sbjct 168 EIQDLEIDHEAELLVLASDGLWD 190
> 7291977
Length=352
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 59/115 (51%), Gaps = 7/115 (6%)
Query 48 VPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRI 107
+ E L+ AN GDSR + G +SVD KPND E R+ +GG V R+
Sbjct 124 IRERRLYCANAGDSRAIAC-ISGMVHALSVDHKPNDAKESKRIMASGGWVEFN-----RV 177
Query 108 DGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG-DILVFACDGVFE 161
+G LALSR+ GDF K P +Q V A P + + + ++ ACDG+++
Sbjct 178 NGNLALSRALGDFIYKKNLLKTPEEQIVTAYPDVEVLDITEDLEFVLLACDGIWD 232
> YDL006w
Length=281
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 49/158 (31%), Positives = 79/158 (50%), Gaps = 30/158 (18%)
Query 19 DAALRSDPEIPI----GPGSTGIIGILLAE----------DPAVPEFTLHVANIGDSRLL 64
D+ L D EI G T + +L E D A + L+ AN+GDSR+
Sbjct 95 DSFLAIDEEINTKLVGNSGCTAAVCVLRWELPDSVSDDSMDLAQHQRKLYTANVGDSRI- 153
Query 65 VLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKG 124
VL +G ++ D K +D +EM RV++AGG +M++ R++G LA++RS GD
Sbjct 154 VLFRNGNSIRLTYDHKASDTLEMQRVEQAGGLIMKS-----RVNGMLAVTRSLGDKFFDS 208
Query 125 RTDILPSQQKVVALPHARTIKVKAGD-ILVFACDGVFE 161
VV P ++++ + D L+ ACDG+++
Sbjct 209 L---------VVGSPFTTSVEITSEDKFLILACDGLWD 237
> 7301459
Length=367
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/110 (37%), Positives = 58/110 (52%), Gaps = 7/110 (6%)
Query 53 LHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLA 112
+++ N GDSR V+ +G ++D KP P E R+Q AGGSVM + RI+G LA
Sbjct 128 IYLVNCGDSRA-VISRNGAAVISTIDHKPFSPKEQERIQNAGGSVM-----IKRINGTLA 181
Query 113 LSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAGD-ILVFACDGVFE 161
+SR+FGD+ K P Q V P D +V ACDG+++
Sbjct 182 VSRAFGDYDFKNDGSKSPVDQMVSPEPDIIVCNRSEHDEFIVVACDGIWD 231
> YBL056w
Length=468
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/112 (36%), Positives = 56/112 (50%), Gaps = 8/112 (7%)
Query 53 LHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLA 112
L AN GDSR VL T G MS D KP E +R+ A G V R++G LA
Sbjct 134 LICANSGDSRT-VLSTGGNSKAMSFDHKPTLLSEKSRIVAADGFVEMD-----RVNGNLA 187
Query 113 LSRSFGDFRMKGRTDILPSQQKVVALPH--ARTIKVKAGDILVFACDGVFEA 162
LSR+ GDF K T + P +Q V +P + + ++ ACDG+++
Sbjct 188 LSRAIGDFEFKSNTKLGPHEQVVTCVPDIICHNLNYDEDEFVILACDGIWDC 239
> At5g24940
Length=447
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/141 (34%), Positives = 71/141 (50%), Gaps = 20/141 (14%)
Query 22 LRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKP 81
L+S+ GST IL+ + L VAN+GDSR ++ F +S D KP
Sbjct 114 LKSENSHTRDAGSTASTAILVGD-------RLLVANVGDSRAVICRGGNAF-AVSRDHKP 165
Query 82 NDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHA 141
+ E R++ AGG VM G WR+ G LA+SR+FGD +K Q VVA P
Sbjct 166 DQSDERERIENAGGFVM--WAGTWRVGGVLAVSRAFGDRLLK---------QYVVADPEI 214
Query 142 RTIKVKAG-DILVFACDGVFE 161
+ K+ + L+ A DG+++
Sbjct 215 QEEKIDDSLEFLILASDGLWD 235
> At4g31860
Length=357
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 44/132 (33%), Positives = 70/132 (53%), Gaps = 16/132 (12%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST + + V + L VAN GDSR ++ + + +S D KP+ E R+ +
Sbjct 160 GSTACVAV-------VRDKQLFVANAGDSRCVISRKNQAYN-LSRDHKPDLEAEKERILK 211
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVV-ALPHARTIKV-KAGD 150
AGG + R++G L LSR+ GD K + LPS++++V A P T+++ D
Sbjct 212 AGGFIH-----AGRVNGSLNLSRAIGDMEFK-QNKFLPSEKQIVTASPDVNTVELCDDDD 265
Query 151 ILVFACDGVFEA 162
LV ACDG+++
Sbjct 266 FLVLACDGIWDC 277
> At4g26080
Length=434
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 55/152 (36%), Positives = 77/152 (50%), Gaps = 30/152 (19%)
Query 18 LDAALRSDPEI----PIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFT 73
++ LR D EI P GST ++ ++ + VAN GDSR VL T
Sbjct 222 FNSFLRVDSEIESVAPETVGSTSVVAVVFPSH-------IFVANCGDSRA-VLCRGKTAL 273
Query 74 PMSVDQKPNDPVEMARVQRAGGSVMRTAMGVW---RIDGRLALSRSFGDFRMKGRTDILP 130
P+SVD KP+ E AR++ AGG V++ W R+ G LA+SRS GD +K P
Sbjct 274 PLSVDHKPDREDEAARIEAAGGKVIQ-----WNGARVFGVLAMSRSIGDRYLK------P 322
Query 131 SQQKVVALPHARTIK-VKAGDILVFACDGVFE 161
S ++ P +K VK D L+ A DGV++
Sbjct 323 S---IIPDPEVTAVKRVKEDDCLILASDGVWD 351
> CE24009
Length=356
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 53/163 (32%), Positives = 78/163 (47%), Gaps = 19/163 (11%)
Query 5 DFSGNSFAEAS----FALDAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGD 60
+FS + EA LD +R D E T + +L+ E ++ N GD
Sbjct 83 EFSEGNMKEAIEKGFLELDQQMRVDEETKDDVSGTTAVVVLIKEG------DVYCGNAGD 136
Query 61 SRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDF 120
SR V G P+S D KP+ E R+ AGG V R++G LALSR+ GDF
Sbjct 137 SRA-VSSVVGEARPLSFDHKPSHETEARRIIAAGGWVEFN-----RVNGNLALSRALGDF 190
Query 121 RMKGRTDILPSQQKVV-ALPHARTIKVKAG-DILVFACDGVFE 161
K D P+++++V A P T K+ + +V ACDG+++
Sbjct 191 AFK-NCDTKPAEEQIVTAFPDVITDKLTPDHEFIVLACDGIWD 232
> 7298474
Length=428
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 81/161 (50%), Gaps = 27/161 (16%)
Query 4 PDFSGNSFAEASFALDAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRL 63
PDF N+F A D + I G+T + ++ + L++A +GDS+
Sbjct 234 PDFYRNAFESAFLLADERF---TQKKITSGTTSVCALITKDQ-------LYIAWVGDSKA 283
Query 64 LVLHTDGTFTPMSV--DQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFR 121
L++ G T + + KP +P E R++ AGG+V+ A G WR++G L ++RS GD+
Sbjct 284 LLV---GKRTQLQLVKPHKPENPDERKRIETAGGTVLH-AQGQWRVNGILNVARSIGDYS 339
Query 122 MKGRTDILPSQQKVVALPHARTIKV-KAGDILVFACDGVFE 161
++ V+A P +++ +A D LV DG+++
Sbjct 340 LEA----------VIAEPDFVDVQLNEAHDFLVLGTDGLWD 370
> At2g25070
Length=355
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 48/134 (35%), Positives = 67/134 (50%), Gaps = 14/134 (10%)
Query 31 GPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARV 90
GP S + L +D L VAN GDSR ++ + +S D KP+ VE R+
Sbjct 156 GPTSGCTACVALIKDKK-----LFVANAGDSRCVISRKSQAYN-LSKDHKPDLEVEKERI 209
Query 91 QRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQ-QKVVALPHARTIKV-KA 148
+AGG + RI+G L L+R+ GD K + LPS+ Q V A P TI +
Sbjct 210 LKAGGFIH-----AGRINGSLNLTRAIGDMEFK-QNKFLPSEKQMVTADPDINTIDLCDD 263
Query 149 GDILVFACDGVFEA 162
D LV ACDG+++
Sbjct 264 DDFLVVACDGIWDC 277
> YER089c
Length=464
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 45/146 (30%), Positives = 67/146 (45%), Gaps = 12/146 (8%)
Query 19 DAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVD 78
D L DP + IL+++ + L N GDSR VL TDG +S D
Sbjct 104 DVKLLQDPVMKEDHSGCTATSILVSKSQNL----LVCGNAGDSRT-VLATDGNAKALSYD 158
Query 79 QKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVAL 138
KP E +R+ A G V R++G LALSR+ GDF K + P +Q V +
Sbjct 159 HKPTLASEKSRIVAADGFVEMD-----RVNGNLALSRAIGDFEFKSNPKLGPEEQIVTCV 213
Query 139 PH--ARTIKVKAGDILVFACDGVFEA 162
P ++ + ++ ACDG+++
Sbjct 214 PDILEHSLDYDRDEFVILACDGIWDC 239
> YBR125c
Length=393
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 50/168 (29%), Positives = 76/168 (45%), Gaps = 36/168 (21%)
Query 19 DAALRSDPEI-----PIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFT 73
DA + D E+ GST ++ ++ E+ +L+VAN GDSR ++
Sbjct 158 DAFILQDEELYRHFANSSCGSTAVVACIINEE------SLYVANCGDSRCILSSKSNGIK 211
Query 74 PMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMK-------GRT 126
MS D KP E+ R+ GG+V ++G R+ G LALSR+F DF+ K RT
Sbjct 212 TMSFDHKPQHIGELIRINDNGGTV---SLG--RVGGVLALSRAFSDFQFKRGVTYPHRRT 266
Query 127 DIL-----------PSQQKVVALPHA--RTIKVKAGDILVFACDGVFE 161
+ P + +V P I + LV ACDG+++
Sbjct 267 KLTNITQNLTYGTPPQEAQVTVEPDVLMHKIDYSKDEFLVLACDGIWD 314
> At2g40180
Length=390
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 59/110 (53%), Gaps = 12/110 (10%)
Query 53 LHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLA 112
L V+N GD R V+ GT ++ D P+ E+ R++ GG V GVWRI G LA
Sbjct 237 LAVSNAGDCRA-VMSRGGTAEALTSDHNPSQANELKRIEALGGYV-DCCNGVWRIQGTLA 294
Query 113 LSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG-DILVFACDGVFE 161
+SR GD +K + V+A P RT+++K + L+ A DG+++
Sbjct 295 VSRGIGDRYLK---------EWVIAEPETRTLRIKPEFEFLILASDGLWD 335
> 7297303
Length=524
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 43/131 (32%), Positives = 64/131 (48%), Gaps = 18/131 (13%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
G+T +I I+ L VAN+GDSR ++ G P+S D KP E R+
Sbjct 323 GTTALIAIVQGS-------KLIVANVGDSRGVMYDWRGIAIPLSFDHKPQQVRERKRIHD 375
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVK--AGD 150
AGG + GVWR+ G LA SR+ GD+ +K + V+A P T ++
Sbjct 376 AGGFI--AFRGVWRVAGVLATSRALGDYPLK-------DKNLVIATPDILTFELNDHKPH 426
Query 151 ILVFACDGVFE 161
L+ A DG+++
Sbjct 427 FLILASDGLWD 437
> At1g43900
Length=369
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 66/133 (49%), Gaps = 21/133 (15%)
Query 29 PIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMA 88
P GST L+ + L VAN+GDSR+ V +G+ P+S D KP+ E
Sbjct 211 PKNAGSTAATAFLIGD-------KLIVANVGDSRV-VASRNGSAVPLSDDHKPDRSDERQ 262
Query 89 RVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKA 148
R++ AGG ++ G WR+ G LA+SR+FGD ++K P Q + L
Sbjct 263 RIEDAGGFIIWA--GTWRVGGILAVSRAFGDKQLKPYVIAEPEIQDISTL---------- 310
Query 149 GDILVFACDGVFE 161
+ +V A DG++
Sbjct 311 -EFIVVASDGLWN 322
> CE05722
Length=468
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 39/109 (35%), Positives = 59/109 (54%), Gaps = 9/109 (8%)
Query 55 VANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALS 114
+ N+GDSR +V + F + D KP E R++ AGGSVM + RI+G LA+S
Sbjct 232 IGNLGDSRAVVAGKNEIFG--TEDHKPYLEKERKRIEGAGGSVM-----IQRINGSLAVS 284
Query 115 RSFGDFRMKGRTDILPSQQKVVALP--HARTIKVKAGDILVFACDGVFE 161
R+FGD+ K + QQ V P + R ++ +V ACDG+++
Sbjct 285 RAFGDYEYKDDPRLPADQQLVSPEPDVYIRERNLENDQFMVVACDGIYD 333
> At1g78200
Length=238
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 43/127 (33%), Positives = 61/127 (48%), Gaps = 15/127 (11%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST + IL + L +AN+GDSR +V + G MSVD P+D E + ++
Sbjct 80 GSTAVTAIL------INGKALWIANVGDSRAIV-SSRGKAKQMSVDHDPDDDTERSMIES 132
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAGDIL 152
GG V V R++G LA+SR FGD +K + P + V H D L
Sbjct 133 KGGFVTNRPGDVPRVNGLLAVSRVFGDKNLKAYLNSEPEIKDVTIDSHT--------DFL 184
Query 153 VFACDGV 159
+ A DG+
Sbjct 185 ILASDGI 191
> At3g11410
Length=399
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 71/132 (53%), Gaps = 24/132 (18%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST ++ ++ PE + V+N GDSR VL +G P+SVD KP+ P E+ R+Q+
Sbjct 221 GSTAVVSVV------TPEKII-VSNCGDSRA-VLCRNGVAIPLSVDHKPDRPDELIRIQQ 272
Query 93 AGGSVMRTAMGVW---RIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG 149
AGG V+ W R+ G LA+SR+ GD +K ++P + V T +
Sbjct 273 AGGRVI-----YWDGARVLGVLAMSRAIGDNYLKPY--VIPDPEVTV------TDRTDED 319
Query 150 DILVFACDGVFE 161
+ L+ A DG+++
Sbjct 320 ECLILASDGLWD 331
> At1g07430
Length=442
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 70/132 (53%), Gaps = 24/132 (18%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST ++ ++ PE + VAN GDSR VL +G P+S D KP+ P E+ R+Q
Sbjct 233 GSTAVVSVI------TPEKII-VANCGDSRA-VLCRNGKAVPLSTDHKPDRPDELDRIQE 284
Query 93 AGGSVMRTAMGVW---RIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG 149
AGG V+ W R+ G LA+SR+ GD +K + S+ +V T + +
Sbjct 285 AGGRVI-----YWDGARVLGVLAMSRAIGDNYLKP---YVTSEPEVTV-----TDRTEED 331
Query 150 DILVFACDGVFE 161
+ L+ A DG+++
Sbjct 332 EFLILATDGLWD 343
> At5g57050
Length=423
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 55/159 (34%), Positives = 79/159 (49%), Gaps = 34/159 (21%)
Query 9 NSFAEASFALDAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLH--VANIGDSRLLVL 66
NSF +D+ + + P GST ++ ++ F H VAN GDSR VL
Sbjct 211 NSF----MRVDSEIETVAHAPETVGSTSVVAVV---------FPTHIFVANCGDSRA-VL 256
Query 67 HTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVW---RIDGRLALSRSFGDFRMK 123
T +SVD KP+ E AR++ AGG V+R W R+ G LA+SRS GD +K
Sbjct 257 CRGKTPLALSVDHKPDRDDEAARIEAAGGKVIR-----WNGARVFGVLAMSRSIGDRYLK 311
Query 124 GRTDILPSQQKVVALPHARTI-KVKAGDILVFACDGVFE 161
PS V+ P ++ +VK D L+ A DG+++
Sbjct 312 ------PS---VIPDPEVTSVRRVKEDDCLILASDGLWD 341
> 7301795
Length=368
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/110 (36%), Positives = 57/110 (51%), Gaps = 7/110 (6%)
Query 53 LHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLA 112
+++AN GDSR VL G + D KP P E R+ AGGSVM + R++G LA
Sbjct 133 VYIANCGDSRA-VLCRQGVPVFATQDHKPILPEEKERIYNAGGSVM-----IKRVNGTLA 186
Query 113 LSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAGD-ILVFACDGVFE 161
+SR+ GD+ K + +Q V P + D LV ACDG+++
Sbjct 187 VSRALGDYDFKNVKEKGQCEQLVSPEPEIFCQSRQDSDEFLVLACDGIWD 236
> At4g31750
Length=389
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 49/147 (33%), Positives = 70/147 (47%), Gaps = 23/147 (15%)
Query 2 RMPDFSGNSFAEASFALDAA----LRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVAN 57
R P F ++ A + A + L+S+ GST IL+ + L VAN
Sbjct 115 RHPKFISDTTAAIADAYNQTDSEFLKSENSQNRDAGSTASTAILVGD-------RLLVAN 167
Query 58 IGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSF 117
+GDSR ++ G +S D KP+ E R++ AGG VM G WR+ G LA+SR+F
Sbjct 168 VGDSRAVICR-GGNAIAVSRDHKPDQSDERQRIEDAGGFVMWA--GTWRVGGVLAVSRAF 224
Query 118 GDFRMKGRTDILPSQQKVVALPHARTI 144
GD +K Q VVA P + +
Sbjct 225 GDRLLK---------QYVVADPEIQVL 242
> Hs20535993
Length=372
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 72/149 (48%), Gaps = 18/149 (12%)
Query 13 EASFALDAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTF 72
+ +F+ A L +D + + G+T + +L L VA++GDSR +L G
Sbjct 168 DKAFSSHARLSADATL-LTSGTTATVALLR------DGIELVVASVGDSRA-ILCRKGKP 219
Query 73 TPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQ 132
+++D P E R+++ GG V ++G ++GRLA++RS GD +K
Sbjct 220 MKLTIDHTPERKDEKERIKKCGGFVAWNSLGQPHVNGRLAMTRSIGDLDLK--------T 271
Query 133 QKVVALPHARTIKVKAGD--ILVFACDGV 159
V+A P + IK+ D LV DG+
Sbjct 272 SGVIAEPETKRIKLHHADDSFLVLTTDGI 300
> At5g51760
Length=416
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 47/138 (34%), Positives = 69/138 (50%), Gaps = 20/138 (14%)
Query 25 DPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDP 84
DP GST + +L + + VAN GDSR VL +G P+S D KP+ P
Sbjct 223 DPREAAISGSTAVTAVLTHDH-------IIVANTGDSRA-VLCRNGMAIPLSNDHKPDRP 274
Query 85 VEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTI 144
E AR++ AGG V+ + R++G LA SR+ GD +K V P +
Sbjct 275 DERARIEAAGGRVL--VVDGARVEGILATSRAIGDRYLK---------PMVAWEPEVTFM 323
Query 145 KVKAGD-ILVFACDGVFE 161
+ ++GD LV A DG+++
Sbjct 324 RRESGDECLVLASDGLWD 341
> At5g59220
Length=413
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 66/133 (49%), Gaps = 26/133 (19%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST ++ +L E + VAN GDSR VL +G +S D KP+ P E+ R+Q
Sbjct 221 GSTAVVSVLTPE-------KIIVANCGDSRA-VLCRNGKAIALSSDHKPDRPDELDRIQA 272
Query 93 AGGSVMRTAMGVW---RIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHAR-TIKVKA 148
AGG V+ W R+ G LA+SR+ GD +K V++ P T +
Sbjct 273 AGGRVI-----YWDGPRVLGVLAMSRAIGDNYLK---------PYVISRPEVTVTDRANG 318
Query 149 GDILVFACDGVFE 161
D L+ A DG+++
Sbjct 319 DDFLILASDGLWD 331
> At2g30020
Length=396
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 60/115 (52%), Gaps = 12/115 (10%)
Query 48 VPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRI 107
V E L V+N GD R V+ G +S D +P+ E R++ GG V T GVWRI
Sbjct 239 VNEGNLVVSNAGDCRA-VMSVGGVAKALSSDHRPSRDDERKRIETTGGYV-DTFHGVWRI 296
Query 108 DGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG-DILVFACDGVFE 161
G LA+SR GD ++K + V+A P + +++ + L+ A DG+++
Sbjct 297 QGSLAVSRGIGDAQLK---------KWVIAEPETKISRIEHDHEFLILASDGLWD 342
> At2g29380
Length=362
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 64/132 (48%), Gaps = 25/132 (18%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST ++ ++ + + VAN GDSR VL +G P+S D KP+ P E+ R++
Sbjct 190 GSTAVVSVITPD-------KIVVANCGDSRA-VLCRNGKPVPLSTDHKPDRPDELDRIEG 241
Query 93 AGGSVMRTAMGVW---RIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG 149
AGG V+ W R+ G LA+SR+ GD +K V P +
Sbjct 242 AGGRVI-----YWDCPRVLGVLAMSRAIGDNYLK---------PYVSCEPEVTITDRRDD 287
Query 150 DILVFACDGVFE 161
D L+ A DG+++
Sbjct 288 DCLILASDGLWD 299
> At3g15260
Length=289
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 52/91 (57%), Gaps = 9/91 (9%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST + IL+ V VAN+GDSR ++ +G P+SVD +PN +E ++
Sbjct 133 GSTAVTAILINCQKLV------VANVGDSRAVICQ-NGVAKPLSVDHEPN--MEKDEIEN 183
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMK 123
GG V V R+DG+LA++R+FGD +K
Sbjct 184 RGGFVSNFPGDVPRVDGQLAVARAFGDKSLK 214
> At1g22280
Length=294
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 50/153 (32%), Positives = 76/153 (49%), Gaps = 19/153 (12%)
Query 10 SFAEASFALDAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTD 69
S A+A D A+ S+ GST + IL + L +AN+GDSR ++ H
Sbjct 115 SIAKAYEKTDQAILSNSSDLGRGGSTAVTAIL------INGRKLWIANVGDSRAVLSHG- 167
Query 70 GTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDIL 129
G T MS D +P E + ++ GG V V R++G+LA+SR+FGD KG L
Sbjct 168 GAITQMSTDHEPR--TERSSIEDRGGFVSNLPGDVPRVNGQLAVSRAFGD---KGLKTHL 222
Query 130 PSQQKVVALPHARTIKVKA-GDILVFACDGVFE 161
S+ P + V + D+L+ A DG+++
Sbjct 223 SSE------PDIKEATVDSQTDVLLLASDGIWK 249
> At3g62260
Length=383
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 48/156 (30%), Positives = 74/156 (47%), Gaps = 24/156 (15%)
Query 9 NSFAEASFALDAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHT 68
N+F +A D AL D I G+T + ++ L VAN GD R VL
Sbjct 166 NAFLQA----DLALAEDCSISDSCGTTALTALICGR-------LLMVANAGDCRA-VLCR 213
Query 69 DGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDI 128
G MS D KP + +E RV+ +GG + ++ LA++R+ GD+ +K
Sbjct 214 KGRAIDMSEDHKPINLLERRRVEESGGFITNDGY----LNEVLAVTRALGDWDLK----- 264
Query 129 LP--SQQKVVALPHARTIKVKAGD-ILVFACDGVFE 161
LP SQ +++ P + I + D LV CDG+++
Sbjct 265 LPHGSQSPLISEPEIKQITLTEDDEFLVIGCDGIWD 300
> At1g72770
Length=511
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 43/132 (32%), Positives = 70/132 (53%), Gaps = 24/132 (18%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST ++ ++ + + V+N GDSR VL P+SVD KP+ E AR++
Sbjct 326 GSTAVVALVCSSH-------IVVSNCGDSRA-VLFRGKEAMPLSVDHKPDREDEYARIEN 377
Query 93 AGGSVMRTAMGVW---RIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAG 149
AGG V++ W R+ G LA+SRS GD +K ++P + +V +P +R
Sbjct 378 AGGKVIQ-----WQGARVFGVLAMSRSIGDRYLKPY--VIP-EPEVTFMPRSR-----ED 424
Query 150 DILVFACDGVFE 161
+ L+ A DG+++
Sbjct 425 ECLILASDGLWD 436
> At2g20630
Length=290
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 64/116 (55%), Gaps = 11/116 (9%)
Query 9 NSFAEASFALDAALRSDPEIPIGPG-STGIIGILLAEDPAVPEFTLHVANIGDSRLLVLH 67
N+ A + DA + + + +G G ST + GIL + TL +AN+GDSR V+
Sbjct 99 NAIRNAYISTDAVIL-EQSLKLGKGGSTAVTGIL------IDGKTLVIANVGDSRA-VMS 150
Query 68 TDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMK 123
+G + +SVD +P+ E ++ GG V V R+DG+LA++R+FGD +K
Sbjct 151 KNGVASQLSVDHEPSK--EQKEIESRGGFVSNIPGDVPRVDGQLAVARAFGDKSLK 204
> At2g25620
Length=392
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 51/159 (32%), Positives = 74/159 (46%), Gaps = 33/159 (20%)
Query 10 SFAEASFALDAALRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTD 69
+F EA +LD +L S G+T + IL +L VAN GD R VL
Sbjct 176 AFLEAC-SLDGSLAS--------GTTALAAILFGR-------SLVVANAGDCRA-VLSRQ 218
Query 70 GTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGR---- 125
G MS D KP E R++ +GG V + +G+L ++R+ GDF M+G
Sbjct 219 GKAIEMSRDHKPMSSKERRRIEASGGHVFDGYL-----NGQLNVARALGDFHMEGMKKKK 273
Query 126 --TDILPSQQKVVALPHARTIKVKAGD-ILVFACDGVFE 161
+D P ++A P T K+ D L+ CDGV++
Sbjct 274 DGSDCGP----LIAEPELMTTKLTEEDEFLIIGCDGVWD 308
> At2g34740
Length=239
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 40/134 (29%), Positives = 70/134 (52%), Gaps = 18/134 (13%)
Query 30 IGP--GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEM 87
+GP GST + I++ V VAN+GDSR ++ ++VD +P+ ++
Sbjct 73 VGPRGGSTAVTAIVIDGKKIV------VANVGDSRAILCRESDVVKQITVDHEPDKERDL 126
Query 88 ARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVK 147
V+ GG V + V R+DG+LA++R+FGD +K ++P+ + +A H T
Sbjct 127 --VKSKGGFVSQKPGNVPRVDGQLAMTRAFGDGGLKEHISVIPNIE--IAEIHDDT---- 178
Query 148 AGDILVFACDGVFE 161
L+ A DG+++
Sbjct 179 --KFLILASDGLWK 190
> Hs7661862
Length=454
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 42/144 (29%), Positives = 65/144 (45%), Gaps = 26/144 (18%)
Query 22 LRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKP 81
LR + G+TG+ ++ TLHVA +GDS+++++ G + +P
Sbjct 243 LRKAKRERLQSGTTGVCALIAGA-------TLHVAWLGDSQVILVQ-QGQVVKLMEPHRP 294
Query 82 NDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMK----GRTDILPSQQKVVA 137
E AR++ GG V + M WR++G LA+SR+ GD K G D
Sbjct 295 ERQDEKARIEALGGFV--SHMDCWRVNGTLAVSRAIGDVFQKPYVSGEADA--------- 343
Query 138 LPHARTIKVKAGDILVFACDGVFE 161
A + D L+ ACDG F+
Sbjct 344 ---ASRALTGSEDYLLLACDGFFD 364
> At5g10740
Length=348
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 48/141 (34%), Positives = 68/141 (48%), Gaps = 26/141 (18%)
Query 22 LRSDPEIPIGPGSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKP 81
L+S+ GST IL+ + L VAN+GDSR V+ G +S D KP
Sbjct 114 LKSENSHNRDAGSTASTAILVGD-------RLVVANVGDSRA-VISRGGKAIAVSRDHKP 165
Query 82 NDPVEMARVQRAGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHA 141
+ E R++ AGG VM W G LA+SR+FGD +K Q VVA P
Sbjct 166 DQSDERERIENAGGFVM------WA--GVLAVSRAFGDRLLK---------QYVVADPEI 208
Query 142 RTIKVKAG-DILVFACDGVFE 161
+ K+ + L+ A DG+++
Sbjct 209 QEEKIDDTLEFLILASDGLWD 229
> At4g28400
Length=268
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/129 (31%), Positives = 63/129 (48%), Gaps = 17/129 (13%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
GST + GIL+ V VAN+GDSR V+ +G +SVD +P+ E ++
Sbjct 98 GSTAVTGILIDGKKLV------VANVGDSRA-VMSKNGVAHQLSVDHEPSK--EKKEIES 148
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVKAGDIL 152
GG V V R+DG+LA++R+FGD +K P H + +
Sbjct 149 RGGFVSNIPGDVPRVDGQLAVARAFGDKSLKLHLSSEPDITHQTIDDHT--------EFI 200
Query 153 VFACDGVFE 161
+FA DG+++
Sbjct 201 LFASDGIWK 209
> At1g07160
Length=380
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 56/111 (50%), Gaps = 12/111 (10%)
Query 52 TLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQRAGGSVMRTAMGVWRIDGRL 111
L VAN GD R VL G ++ D +P+ E R++ +GG V T VWRI G L
Sbjct 226 NLVVANAGDCRA-VLSVGGFAEALTSDHRPSRDDERNRIESSGGYV-DTFNSVWRIQGSL 283
Query 112 ALSRSFGDFRMKGRTDILPSQQKVVALPHARTIKVK-AGDILVFACDGVFE 161
A+SR GD +K Q +++ P +++ + L+ A DG+++
Sbjct 284 AVSRGIGDAHLK---------QWIISEPEINILRINPQHEFLILASDGLWD 325
> HsM7662472
Length=757
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 38/133 (28%), Positives = 62/133 (46%), Gaps = 26/133 (19%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
G+TG++ + LHVA +GDS+++++ G + KP+ E R++
Sbjct 331 GTTGVVTFIRGN-------MLHVAWVGDSQVMLVRK-GQAVELMKPHKPDREDEKQRIEA 382
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMK----GRTDILPSQQKVVALPHARTIKVKA 148
GG V+ G WR++G L++SR+ GD K G D A T+
Sbjct 383 LGGCVV--WFGAWRVNGSLSVSRAIGDAEHKPYICGDAD------------SASTVLDGT 428
Query 149 GDILVFACDGVFE 161
D L+ ACDG ++
Sbjct 429 EDYLILACDGFYD 441
> Hs21361464
Length=766
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 37/133 (27%), Positives = 62/133 (46%), Gaps = 26/133 (19%)
Query 33 GSTGIIGILLAEDPAVPEFTLHVANIGDSRLLVLHTDGTFTPMSVDQKPNDPVEMARVQR 92
G+TG++ + LHVA +GDS+++++ G + KP+ E R++
Sbjct 340 GTTGVVTFIRGN-------MLHVAWVGDSQVMLVRK-GQAVELMKPHKPDREDEKQRIEA 391
Query 93 AGGSVMRTAMGVWRIDGRLALSRSFGDFRMK----GRTDILPSQQKVVALPHARTIKVKA 148
GG ++ G WR++G L++SR+ GD K G D A T+
Sbjct 392 LGGCIV--WFGAWRVNGSLSVSRAIGDAEHKPYICGDAD------------SASTVLDGT 437
Query 149 GDILVFACDGVFE 161
D L+ ACDG ++
Sbjct 438 EDYLILACDGFYD 450
Lambda K H
0.321 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2317343104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40