bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
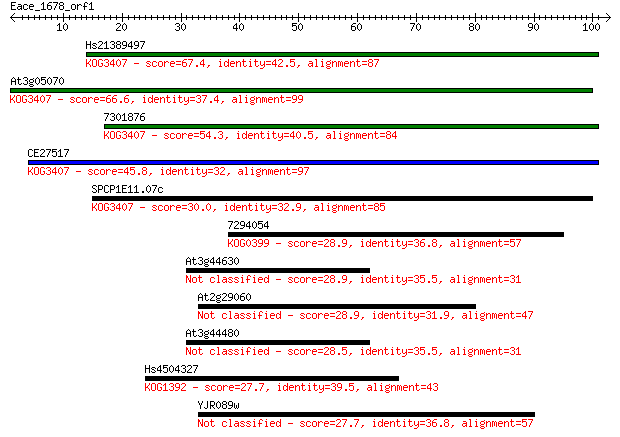
Query= Eace_1678_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
Hs21389497 67.4 6e-12
At3g05070 66.6 1e-11
7301876 54.3 6e-08
CE27517 45.8 2e-05
SPCP1E11.07c 30.0 1.1
7294054 28.9 2.4
At3g44630 28.9 2.7
At2g29060 28.9 2.7
At3g44480 28.5 2.8
Hs4504327 27.7 5.0
YJR089w 27.7 5.0
> Hs21389497
Length=166
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 61/92 (66%), Gaps = 9/92 (9%)
Query 14 ELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDI-----LSQVVPRR 68
ELR RNYVP+D L++ +P+ +E+++ + ++AAK E + L+ + PR+
Sbjct 56 ELRLRNYVPEDEDLKKRRVPQAKPVAVEEKV----KEQLEAAKPEPVIEEVDLANLAPRK 111
Query 69 PNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 112 PDWDLKRDVAKKLEKLKKRTQRAIAELIRERL 143
> At3g05070
Length=144
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 37/99 (37%), Positives = 60/99 (60%), Gaps = 3/99 (3%)
Query 1 EDVEESSSLSSSFELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDI 60
ED E+ + ++FRNYVP+ ++L+ L P + + E I + AV+ K ED
Sbjct 33 EDGEDEAVEEDGPAMKFRNYVPQAKELQDGKLAPPELPKFEDPI-VALPPAVE--KKEDP 89
Query 61 LSQVVPRRPNWDLKRDVERKIAILSRRTDKAIIQLIREK 99
+ P++PNWDL+RDV++K+ L RRT KA+ +L+ E+
Sbjct 90 FVNIAPKKPNWDLRRDVQKKLDKLERRTQKAMHKLMEEQ 128
> 7301876
Length=161
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 53/88 (60%), Gaps = 7/88 (7%)
Query 17 FRNYVPKDRKLRQFCL---PRPSIDE-LEKQIALEAESAVQAAKDEDILSQVVPRRPNWD 72
FR+Y P++ L P +I+ +E Q++L + V DE ++ + PR+P+WD
Sbjct 49 FRSYKPQNETTDGEILAQEPTGNIESAVEDQLSLLQQPMV---IDEIDITNLAPRKPDWD 105
Query 73 LKRDVERKIAILSRRTDKAIIQLIREKI 100
LKRD +K+ L RRT KAI +LIRE++
Sbjct 106 LKRDASKKLERLERRTQKAIAELIRERL 133
> CE27517
Length=169
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 52/98 (53%), Gaps = 3/98 (3%)
Query 4 EESSSLSSSFELRFRNYVPKDRKLRQFCLPRPSIDELEKQIALEAESAVQ-AAKDEDILS 62
E S+ S FRN+ P D Q +D ++++I + + A D L+
Sbjct 55 ETSTKKSREVGREFRNHKPDDAVGTQNV--DMDLDIVQREITEHLKDVLHEKAIDSVDLA 112
Query 63 QVVPRRPNWDLKRDVERKIAILSRRTDKAIIQLIREKI 100
+ P++ +WDLKRD+E K+ L RRT KA+ +IR+++
Sbjct 113 MLAPKKIDWDLKRDIESKLQKLERRTQKAVATIIRQRL 150
> SPCP1E11.07c
Length=142
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 50/99 (50%), Gaps = 16/99 (16%)
Query 15 LRFRNYVPKDRKLRQFCLPRP----SIDELEKQI------ALEAESAVQAAKDEDILSQV 64
+ RNY P+ + + + P S++ L K+I +E +S+V +E L +
Sbjct 39 IEHRNYDPEVQAPKMGFVEPPNMIESVEALSKEIEEKTKRKIEEQSSVPV--EELDLVTL 96
Query 65 VPRRPNWDLKRDVERKIAILSRRTDKAII----QLIREK 99
P++P WDL+RD++ ++ L + AI QLI E+
Sbjct 97 RPKKPTWDLERDLKERMRSLETQNQNAIAFYIQQLISER 135
> 7294054
Length=2162
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 11/68 (16%)
Query 38 DELEKQIALEA---------ESA--VQAAKDEDILSQVVPRRPNWDLKRDVERKIAILSR 86
D+L K + LE +SA V A K E + QV RRP ++ ER++ +L +
Sbjct 179 DDLAKSLGLEVIAWRTVPSNQSAIGVVARKSEPLSRQVFVRRPAGSDEKAFERQVFVLRK 238
Query 87 RTDKAIIQ 94
R +I+
Sbjct 239 RASHELIK 246
> At3g44630
Length=1214
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 16/31 (51%), Gaps = 0/31 (0%)
Query 31 CLPRPSIDELEKQIALEAESAVQAAKDEDIL 61
C PRP DE Q+ L+ + Q +DI+
Sbjct 329 CYPRPCFDEYSAQLQLQNQMLSQMINHKDIM 359
> At2g29060
Length=1336
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 33 PRPSIDELEKQIALEAESAVQAAKDEDILSQVVPRRPNWDLKRDVER 79
P+P + + K + +AESA+Q K + S+ +P W + D+ER
Sbjct 815 PQPVNEIMVKSMFSDAESALQFKKGVEEASKFLPNSDQWVINLDIER 861
> At3g44480
Length=1220
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 16/31 (51%), Gaps = 0/31 (0%)
Query 31 CLPRPSIDELEKQIALEAESAVQAAKDEDIL 61
C PRP DE Q+ L+ + Q +DI+
Sbjct 333 CYPRPCFDEYSAQLQLQNQMLSQMINHKDIM 363
> Hs4504327
Length=474
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 22/43 (51%), Gaps = 4/43 (9%)
Query 24 DRKLRQFCLPRPSIDELEKQIALEAESAVQAAKDEDILSQVVP 66
DR F + R DE AL + S + A+DE +LS VVP
Sbjct 228 DRLAAAFAVSRLEQDEY----ALRSHSLAKKAQDEGLLSDVVP 266
> YJR089w
Length=954
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 29/61 (47%), Gaps = 11/61 (18%)
Query 33 PRPSIDELEKQIALEAESAVQAAKDEDIL----SQVVPRRPNWDLKRDVERKIAILSRRT 88
PR DE + + +L SA KD+D++ S ++ RDV RK AIL T
Sbjct 396 PRKIFDEEDSEHSLNNNSANGDNKDKDLVIDFTSHIIKN-------RDVGRKNAILDDST 448
Query 89 D 89
D
Sbjct 449 D 449
Lambda K H
0.316 0.132 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181107380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40