bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1691_orf1
Length=91
Score E
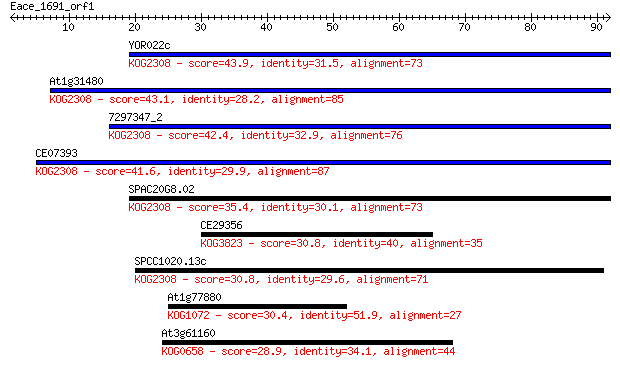
Sequences producing significant alignments: (Bits) Value
YOR022c 43.9 7e-05
At1g31480 43.1 1e-04
7297347_2 42.4 2e-04
CE07393 41.6 4e-04
SPAC20G8.02 35.4 0.023
CE29356 30.8 0.58
SPCC1020.13c 30.8 0.72
At1g77880 30.4 0.91
At3g61160 28.9 2.2
> YOR022c
Length=715
Score = 43.9 bits (102), Expect = 7e-05, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 42/74 (56%), Gaps = 4/74 (5%)
Query 19 FQSIGISSLYEVRRLLSLTAADLLFFMTPRFGDFIMQQVAQQLNAAFRRLKQ-HPSGRFA 77
+ ++ + +R+LL+ D+L ++ P + D I+QQV QLN +R K+ +P
Sbjct 435 LSQVTVNGVLPLRKLLADGLLDILLYVEPYYQDMILQQVTSQLNKTYRIFKEFNPE---F 491
Query 78 KSQFSLLGYSLGSV 91
+ L+G+SLGS+
Sbjct 492 DGKVHLVGHSLGSM 505
> At1g31480
Length=869
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/86 (27%), Positives = 47/86 (54%), Gaps = 4/86 (4%)
Query 7 WKRCIVEAQDHLFQSIGISSLYEVRRLLSLTAADLLFFMTPRFGDFIMQQVAQQLNAAFR 66
W++ + + + + + R +LS T D+L++M+P + I+ V++QLN +
Sbjct 366 WRKGLKLSGEAAVDKCTLDGVRRFREMLSATVHDVLYYMSPIYCQAIIDSVSKQLNRLYL 425
Query 67 R-LKQHPSGRFAKSQFSLLGYSLGSV 91
+ LK++P + S+ G+SLGSV
Sbjct 426 KFLKRNPD---YVGKISIYGHSLGSV 448
> 7297347_2
Length=707
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 25/77 (32%), Positives = 41/77 (53%), Gaps = 4/77 (5%)
Query 16 DHLFQSIGISSLYEVRRLLSLTAADLLFFMTPRFGDFIMQQVAQQLNAAFRRLK-QHPSG 74
D +SI + S+ +R + T D+LF+ +P++ IM VA LN + + + +HP
Sbjct 306 DEKLKSITLESIPRLRNFTNDTLLDVLFYTSPKYCQKIMNTVADALNDVYLKYRMRHPE- 364
Query 75 RFAKSQFSLLGYSLGSV 91
SL G+SLGS+
Sbjct 365 --FNGGVSLAGHSLGSL 379
> CE07393
Length=765
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 48/87 (55%), Gaps = 3/87 (3%)
Query 5 INWKRCIVEAQDHLFQSIGISSLYEVRRLLSLTAADLLFFMTPRFGDFIMQQVAQQLNAA 64
+ W R ++ + L +I I + +R L+ TA D++++ +P F I++ V QLN
Sbjct 335 VEW-RSALKLDNGLTDNITIPKMSSMRASLNSTAMDVMYYQSPLFRTEIVRGVVSQLNRT 393
Query 65 FRRLKQHPSGRFAKSQFSLLGYSLGSV 91
++ K + + +F S+ G+SLGSV
Sbjct 394 YKLFKAN-NPQF-NGHVSVFGHSLGSV 418
> SPAC20G8.02
Length=757
Score = 35.4 bits (80), Expect = 0.023, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 19 FQSIGISSLYEVRRLLSLTAADLLFFMTPRFGDFIMQQVAQQLNAAFRRLKQH-PSGRFA 77
+I I ++ +R ++S D+L + P + D I+ V ++LN + K++ PS
Sbjct 458 LDNINIPTVTGLRNIISDVLLDVLLYCQPNYRDKILAAVVKRLNRLYNLYKKNVPS---F 514
Query 78 KSQFSLLGYSLGSV 91
SLLG+SLG++
Sbjct 515 NGHVSLLGHSLGAL 528
> CE29356
Length=530
Score = 30.8 bits (68), Expect = 0.58, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 24/35 (68%), Gaps = 6/35 (17%)
Query 30 VRRLLSLTAADLLFFMTPRFGDFIMQQVAQQLNAA 64
+ R++SLT ADL RF DFI+++V +Q+ ++
Sbjct 357 LERVVSLTTADL------RFADFILKKVEEQVKSS 385
> SPCC1020.13c
Length=669
Score = 30.8 bits (68), Expect = 0.72, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 39/72 (54%), Gaps = 2/72 (2%)
Query 20 QSIGISSLYEVRRLLSLTAADLLFFMTPRFGDFIMQQVAQQLNAAFRRLKQ-HPSGRFAK 78
+ I I S+ VRRLL +D+ ++M+ + I++ V ++ N + K +P K
Sbjct 333 EDIEIDSIPAVRRLLGDVMSDIPYYMS-HHKESIIKSVIREANRVYHLWKDCNPYFLENK 391
Query 79 SQFSLLGYSLGS 90
+ ++G+SLGS
Sbjct 392 GRIFIIGHSLGS 403
> At1g77880
Length=130
Score = 30.4 bits (67), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 25 SSLYEVRRLLSLTAADLLFFMTPRFGD 51
+ LYE RRLL + + L FFMT R D
Sbjct 58 TELYETRRLLGTSESCLYFFMTERRND 84
> At3g61160
Length=431
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 24 ISSLYEVRRLLSLTAADLLFFMTPRFGDFIMQQV-AQQLNAAFRR 67
+ L E+ ++L A + + M PR+ DF Q+ AQ + FRR
Sbjct 309 VDQLVEIIKILGTPAREEIKNMNPRYNDFKFPQIKAQPWHKIFRR 353
Lambda K H
0.329 0.139 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40