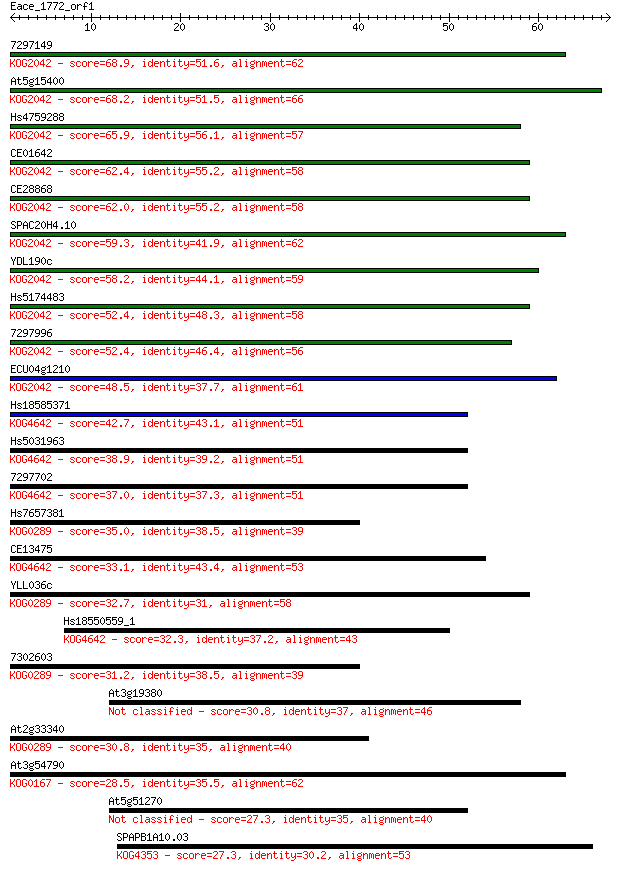
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1772_orf1
Length=67
Score E
Sequences producing significant alignments: (Bits) Value
7297149 68.9 2e-12
At5g15400 68.2 3e-12
Hs4759288 65.9 2e-11
CE01642 62.4 2e-10
CE28868 62.0 2e-10
SPAC20H4.10 59.3 1e-09
YDL190c 58.2 4e-09
Hs5174483 52.4 2e-07
7297996 52.4 2e-07
ECU04g1210 48.5 3e-06
Hs18585371 42.7 1e-04
Hs5031963 38.9 0.002
7297702 37.0 0.008
Hs7657381 35.0 0.031
CE13475 33.1 0.11
YLL036c 32.7 0.16
Hs18550559_1 32.3 0.21
7302603 31.2 0.46
At3g19380 30.8 0.56
At2g33340 30.8 0.66
At3g54790 28.5 2.9
At5g51270 27.3 6.1
SPAPB1A10.03 27.3 6.5
> 7297149
Length=972
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/62 (51%), Positives = 45/62 (72%), Gaps = 0/62 (0%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKAAA 60
PV+LPSS+V VDR I RHL+S+ +DPFNR PLT +++ LK+ IE + + KR+AA
Sbjct 910 PVVLPSSKVTVDRSTIARHLLSDQTDPFNREPLTMDKVKSNEALKQEIESWIQGKREAAR 969
Query 61 AN 62
+N
Sbjct 970 SN 971
> At5g15400
Length=1038
Score = 68.2 bits (165), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 48/69 (69%), Gaps = 3/69 (4%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFT---EAKRK 57
PV+LPSSR+ VDR I+RHL+S+ DPFNRA LT + LIP ELK I+EF ++K++
Sbjct 957 PVILPSSRITVDRPIIQRHLLSDNHDPFNRAHLTSDMLIPDIELKAKIDEFVKSHQSKKR 1016
Query 58 AAAANRPSK 66
+ + +K
Sbjct 1017 TSGEDSSNK 1025
> Hs4759288
Length=1073
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 32/57 (56%), Positives = 44/57 (77%), Gaps = 1/57 (1%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRK 57
PV+LPSSRV VDR I RHL+S+ +DPFNR+PLT +++ P ELK+ I+ + A+RK
Sbjct 1011 PVVLPSSRVTVDRSTIARHLLSDQTDPFNRSPLTMDQIRPNTELKEKIQRWL-AERK 1066
> CE01642
Length=980
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/58 (55%), Positives = 40/58 (68%), Gaps = 1/58 (1%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKA 58
PV LPS V+ DR IERHL+S P++PFNRAPL+ EL P ELK I+E+ KR +
Sbjct 922 PVKLPSGHVM-DRAVIERHLLSTPNNPFNRAPLSHNELSPDSELKAKIQEWICQKRNS 978
> CE28868
Length=984
Score = 62.0 bits (149), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/58 (55%), Positives = 40/58 (68%), Gaps = 1/58 (1%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKA 58
PV LPS V+ DR IERHL+S P++PFNRAPL+ EL P ELK I+E+ KR +
Sbjct 926 PVKLPSGHVM-DRAVIERHLLSTPNNPFNRAPLSHNELSPDSELKAKIQEWICQKRNS 982
> SPAC20H4.10
Length=1010
Score = 59.3 bits (142), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 41/62 (66%), Gaps = 0/62 (0%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKAAA 60
PV+LP S + +DR I+ HL+S+ +DPFNR PLT +++ P L++ I F ++KR +
Sbjct 947 PVVLPRSGISIDRSTIKAHLLSDATDPFNRTPLTLDDVTPNDTLREEINTFLKSKRNKHS 1006
Query 61 AN 62
N
Sbjct 1007 RN 1008
> YDL190c
Length=961
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 42/59 (71%), Gaps = 0/59 (0%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKAA 59
PV+LP+S++ +DR I+ HL+S+ +DPFNR PL E++ P EL++ I F + K++ A
Sbjct 897 PVILPASKMNIDRSTIKAHLLSDSTDPFNRMPLKLEDVTPNEELRQKILCFKKQKKEEA 955
> Hs5174483
Length=1302
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 37/58 (63%), Gaps = 1/58 (1%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKA 58
PV LPS I+DR I RHL++ P+DPFNR LT L P PELK+ I+ + K+ +
Sbjct 1244 PVRLPSG-TIMDRSIILRHLLNSPTDPFNRQTLTESMLEPVPELKEQIQAWMREKQNS 1300
> 7297996
Length=1217
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 38/56 (67%), Gaps = 1/56 (1%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKR 56
PV+LPS V+ DR I RHL++ +DPFNR PLT + L+ ELK+ I+ + + +R
Sbjct 1157 PVVLPSGTVM-DRAIITRHLLNSCTDPFNRQPLTEDMLVANIELKQRIDAWRKEQR 1211
> ECU04g1210
Length=809
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 33/61 (54%), Gaps = 0/61 (0%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKAAA 60
PV L +SR+ VDR + +M+ DPFNR PLT + +I ELK+ I + +
Sbjct 749 PVKLLTSRITVDRSTYDMLMMNGGIDPFNRMPLTEDMVIEDAELKEKINRYYGCSGQGGE 808
Query 61 A 61
A
Sbjct 809 A 809
> Hs18585371
Length=303
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 32/52 (61%), Gaps = 2/52 (3%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPS-DPFNRAPLTREELIPQPELKKAIEEF 51
P + PS + DR++IE HL DP R+PLT+E+LIP +K+ I+ F
Sbjct 243 PCITPSG-ITYDRKDIEEHLQRVGHFDPVTRSPLTQEQLIPNLAMKEVIDAF 293
> Hs5031963
Length=303
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 30/52 (57%), Gaps = 2/52 (3%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPS-DPFNRAPLTREELIPQPELKKAIEEF 51
P + PS + DR++IE HL DP +PLT+E+ IP +K+ I+ F
Sbjct 243 PCITPSG-ITYDRKDIEEHLQRVGHFDPVTGSPLTQEQFIPNLAMKEVIDAF 293
> 7297702
Length=289
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 31/52 (59%), Gaps = 2/52 (3%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPS-DPFNRAPLTREELIPQPELKKAIEEF 51
PV+ PS + +R++IE HL DP R LT+++LIP +K+ ++ F
Sbjct 229 PVITPSG-ITYERKDIEEHLQRVGHFDPVTRVKLTQDQLIPNFSMKEVVDSF 279
> Hs7657381
Length=504
Score = 35.0 bits (79), Expect = 0.031, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELI 39
P + P S + +RR IE+++ +DP N PL+ E+LI
Sbjct 15 PCVSPVSNHVYERRLIEKYIAENGTDPINNQPLSEEQLI 53
> CE13475
Length=272
Score = 33.1 bits (74), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 31/60 (51%), Gaps = 8/60 (13%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPS-DPFNRAPLTREELIPQPELK------KAIEEFTE 53
PV++PS + DR I +HL DP R PLT E+IP LK + IE+F +
Sbjct 201 PVIVPSG-ITYDREEIVQHLRRIGHFDPVTRKPLTENEIIPNYALKEVGNLYRVIEKFLD 259
> YLL036c
Length=503
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 30/61 (49%), Gaps = 3/61 (4%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTRE---ELIPQPELKKAIEEFTEAKRK 57
PV+ P SR I ++ +E+++ +DP PL+ E E++P + E A K
Sbjct 13 PVLSPKSRTIFEKSLLEQYVKDTGNDPITNEPLSIEEIVEIVPSAQQASLTESTNSATLK 72
Query 58 A 58
A
Sbjct 73 A 73
> Hs18550559_1
Length=276
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 7 SRVIVDRRNIERHLM-SEPSDPFNRAPLTREELIPQPELKKAIE 49
+ + DR++IE HL + DP +PLT+++ IP +K+A E
Sbjct 38 TGISYDRKDIEEHLQRVDHFDPVTWSPLTQKQFIPNLAMKEAGE 81
> 7302603
Length=505
Score = 31.2 bits (69), Expect = 0.46, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELI 39
PV+ P S + ++R IE++L+ DP + L EELI
Sbjct 15 PVVSPHSGAVFEKRVIEKYLLENGCDPISGKELKPEELI 53
> At3g19380
Length=428
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 26/48 (54%), Gaps = 2/48 (4%)
Query 12 DRRNIERH--LMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRK 57
DR +IE + + + P RAPL+ LIP L++ I+E+ A R
Sbjct 47 DRASIESWVSIGNNTTCPVTRAPLSDFTLIPNHTLRRLIQEWCVANRS 94
> At2g33340
Length=540
Score = 30.8 bits (68), Expect = 0.66, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSDPFNRAPLTREELIP 40
PV+ S ++ +RR IERH+ P PLT ++++P
Sbjct 13 PVVSTKSGLLFERRLIERHISDYGKCPVTGEPLTIDDIVP 52
> At3g54790
Length=727
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 2/63 (3%)
Query 1 PVMLPSSRVIVDRRNIERHLMSEPSD-PFNRAPLTREELIPQPELKKAIEEFTEAKRKAA 59
PV++ S + DR +I++ L + + P R LT +ELIP +K I + EA R
Sbjct 218 PVIVASGQTF-DRTSIKKWLDNGLAVCPRTRQVLTHQELIPNYTVKAMIASWLEANRINL 276
Query 60 AAN 62
A N
Sbjct 277 ATN 279
> At5g51270
Length=819
Score = 27.3 bits (59), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 12 DRRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEF 51
DRR IE + + + P +PL L+P L AI E+
Sbjct 775 DRRAIEEWMENHRTSPVTNSPLQNVNLLPNHTLYAAIVEW 814
> SPAPB1A10.03
Length=200
Score = 27.3 bits (59), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 27/53 (50%), Gaps = 3/53 (5%)
Query 13 RRNIERHLMSEPSDPFNRAPLTREELIPQPELKKAIEEFTEAKRKAAAANRPS 65
R+ E++ + S P +PLT ++ PE K+ E TE +K+ N P+
Sbjct 143 RQFFEKYGLEAGSKP---SPLTILKITRDPEFKQLAETITEIFKKSGIQNDPA 192
Lambda K H
0.313 0.128 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160559522
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40