bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1774_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
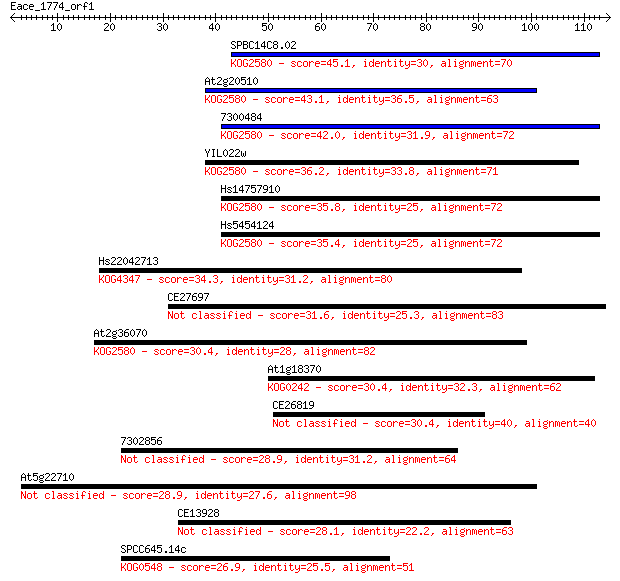
SPBC14C8.02 45.1 3e-05
At2g20510 43.1 1e-04
7300484 42.0 3e-04
YIL022w 36.2 0.017
Hs14757910 35.8 0.022
Hs5454124 35.4 0.024
Hs22042713 34.3 0.053
CE27697 31.6 0.39
At2g36070 30.4 0.77
At1g18370 30.4 0.82
CE26819 30.4 0.83
7302856 28.9 2.3
At5g22710 28.9 2.7
CE13928 28.1 3.6
SPCC645.14c 26.9 8.2
> SPBC14C8.02
Length=427
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 35/70 (50%), Gaps = 0/70 (0%)
Query 43 GQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIGDEETLRLHCGEGA 102
++F ETE + +R K IDPSF + ++ I + AY+ GD+E L+ E
Sbjct 258 SRMFSETEASQVMRRFKEIDPSFNTEHFLQYLREYIVPEVTEAYVKGDKEVLKTWLSEAP 317
Query 103 FAALKASIDE 112
F+ + + E
Sbjct 318 FSVYETTTKE 327
> At2g20510
Length=533
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 44/70 (62%), Gaps = 8/70 (11%)
Query 38 ENPLVGQL-------FKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIGD 90
+NP+V ++ KET+ A++ +E+++ DPSF L + ++ VI + +++AY GD
Sbjct 354 DNPIVHKIQDMNEKFLKETDSASTYKEIRSRDPSFSLPDFAAEIEEVI-KPVLNAYSEGD 412
Query 91 EETLRLHCGE 100
ETL+ +C +
Sbjct 413 VETLKKYCSK 422
> 7300484
Length=459
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 39/72 (54%), Gaps = 0/72 (0%)
Query 41 LVGQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIGDEETLRLHCGE 100
++G LF +TE++ ++ E+ IDPSF + +T I +I+ + + GD E L+ C E
Sbjct 293 VMGGLFSKTELSETMTELVKIDPSFDQKDFLRDCETDIIPNILESIVRGDLEILKDWCFE 352
Query 101 GAFAALKASIDE 112
F + I E
Sbjct 353 STFNIIANPIKE 364
> YIL022w
Length=431
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 34/81 (41%), Gaps = 10/81 (12%)
Query 38 ENPL----------VGQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYL 87
ENPL VG F ETE + + K +DP+F ++ I I+ AY+
Sbjct 247 ENPLIVVMRKITNKVGGFFAETESSRVYSQFKLMDPTFSNESFTRHLREYIVPEILEAYV 306
Query 88 IGDEETLRLHCGEGAFAALKA 108
GD + L+ E F A
Sbjct 307 KGDVKVLKKWFSEAPFNVYAA 327
> Hs14757910
Length=452
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 41 LVGQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIGDEETLRLHCGE 100
L+G LF +TE++ + E+ +DP+F + + I +++ A + G+ + L+ C E
Sbjct 286 LLGGLFSKTEMSEVLTEILRVDPAFDKDRFLKQCENDIIPNVLEAMISGELDILKDWCYE 345
Query 101 GAFAALKASIDE 112
++ L I +
Sbjct 346 ATYSQLAHPIQQ 357
> Hs5454124
Length=452
Score = 35.4 bits (80), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 41 LVGQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIGDEETLRLHCGE 100
L+G LF +TE++ + E+ +DP+F + + I +++ A + G+ + L+ C E
Sbjct 286 LLGGLFSKTEMSEVLTEILRVDPAFDKDRFLKQCENDIIPNVLEAMISGELDILKDWCYE 345
Query 101 GAFAALKASIDE 112
++ L I +
Sbjct 346 ATYSQLAHPIQQ 357
> Hs22042713
Length=1266
Score = 34.3 bits (77), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 1/80 (1%)
Query 18 TAWDKFGSRLRDMPFLQNFFENPLVGQLFKETEIAASIREMKTIDPSFRLSELHELVQTV 77
T+++KFG+ D+ F + V Q ++T +R + T + SF + EL EL
Sbjct 776 TSYEKFGTIRADLIEQMRFKQRLKVIQTLEDTTKRNVVRTIVT-ETSFTIDELEELYALF 834
Query 78 IAEHIISAYLIGDEETLRLH 97
AEH+ S Y G L H
Sbjct 835 KAEHLTSCYWGGSSNALDRH 854
> CE27697
Length=791
Score = 31.6 bits (70), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 21/84 (25%), Positives = 38/84 (45%), Gaps = 4/84 (4%)
Query 31 PFLQNF-FENPLVGQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIG 89
P ++N F N + LFK TE+ + + I+ EL Q+ +AE+ S +
Sbjct 253 PVIENISFNNEIAEPLFKPTELTKNTNPVNEIETPITTEELITETQSTVAENPTSPEVTS 312
Query 90 DEETLRLHCGEGAFAALKASIDER 113
++ LH E + + S+ E+
Sbjct 313 GQD---LHPNEQSAEKPQGSLSEK 333
> At2g36070
Length=499
Score = 30.4 bits (67), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 40/93 (43%), Gaps = 15/93 (16%)
Query 17 ETAWDKFGSRLRDMPFLQNF--FENPLVGQLFKETEIAASIRE---------MKTIDPSF 65
+ W+ +++ P + P+V K EIA +RE + I SF
Sbjct 298 QKKWESLREKMQGYPVFKRLSGMSEPVVN---KSQEIAEDVREKWETSDNPIVHKIQESF 354
Query 66 RLSELHELVQTVIAEHIISAYLIGDEETLRLHC 98
L + +Q I +++AY GD +TL+ +C
Sbjct 355 SLPDFVSEIQEAIRP-VLNAYSKGDAKTLKKYC 386
> At1g18370
Length=954
Score = 30.4 bits (67), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 5/62 (8%)
Query 50 EIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIGDEETLRLHCGEGAFAALKAS 109
++ A IREM+T+ PS L E+ ++ + +SA L EE RLH A L+
Sbjct 514 KLQAEIREMRTVKPSAMLKEVGDV---IAPNKSVSANL--KEEITRLHSQGSTIANLEEQ 568
Query 110 ID 111
++
Sbjct 569 LE 570
> CE26819
Length=498
Score = 30.4 bits (67), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 51 IAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLI-GD 90
+AA++ +K P R +++H L + VI EH+ A LI GD
Sbjct 307 LAANLAVLKAAKPGVRWTDMHILSEKVILEHLKQAGLIVGD 347
> 7302856
Length=1739
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 30/65 (46%), Gaps = 1/65 (1%)
Query 22 KFGSRLRDMPFLQNFFENPLVGQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIA-E 80
+ ++L +LQ + L K E+ + +M T DPSF L E VQ V E
Sbjct 549 RIHTQLLSQSYLQTYSHPTLYPMAKKPREMLEELHKMATKDPSFNCWNLQEAVQLVRKWE 608
Query 81 HIISA 85
H +S+
Sbjct 609 HDLSS 613
> At5g22710
Length=262
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 48/108 (44%), Gaps = 13/108 (12%)
Query 3 LDDKPRETSLVVSSETAWDK-FGSRLRDMPFLQNFF----ENPLVGQLFKETEIAASIRE 57
L P T L V+S + K L+ P L++ F + P+ +K + S+
Sbjct 119 LPQFPHMTRLCVNSYEYYLKMLPMLLKSCPNLKSLFLIYMQEPME---YKNEQTMLSLPV 175
Query 58 MKTIDPSFRLSELHELVQTVIAEHIISAYLIGDEE-----TLRLHCGE 100
+ + PS ++ + AE I++ Y + +E TLRL+CG+
Sbjct 176 PECLRPSLEYIKMKRPINRAGAEMILANYFLENEAVLKKFTLRLYCGK 223
> CE13928
Length=955
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 14/63 (22%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 33 LQNFFENPLVGQLFKETEIAASIREMKTIDPSFRLSELHELVQTVIAEHIISAYLIGDEE 92
++ ENPL+ FK+ + K I+P F + +H+ +Q ++ + I + +E+
Sbjct 174 FNDYLENPLLQLEFKKHTFEELFVQWKQINPLFEMESIHDSLQDLLKKFIARNPIRDNED 233
Query 93 TLR 95
+
Sbjct 234 VFK 236
> SPCC645.14c
Length=591
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 22 KFGSRLRDMPFLQNFFENPLVGQLFKETEIAASIREMKTIDPSFRLSELHE 72
K GS+L D F++ NP L ++ A +++++ +P ++EL++
Sbjct 132 KLGSQLSDPKFMEKLASNPETASLLADSAFMAKLQKIQQ-NPGSIMAELND 181
Lambda K H
0.319 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181971906
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40