bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1816_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
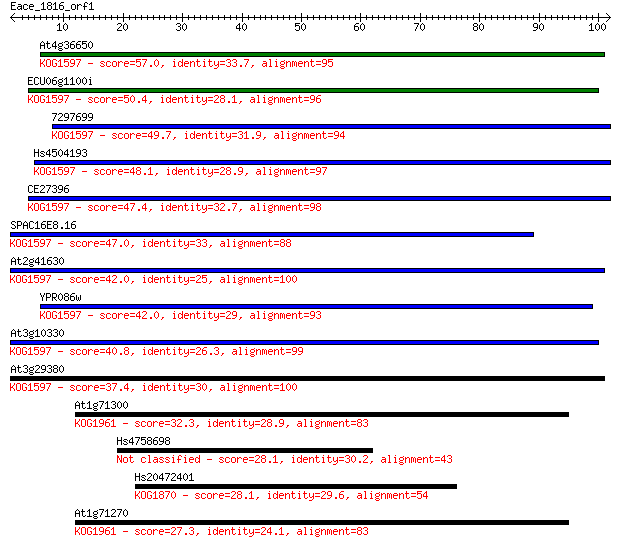
At4g36650 57.0 8e-09
ECU06g1100i 50.4 9e-07
7297699 49.7 1e-06
Hs4504193 48.1 4e-06
CE27396 47.4 6e-06
SPAC16E8.16 47.0 9e-06
At2g41630 42.0 3e-04
YPR086w 42.0 3e-04
At3g10330 40.8 6e-04
At3g29380 37.4 0.007
At1g71300 32.3 0.23
Hs4758698 28.1 4.1
Hs20472401 28.1 4.6
At1g71270 27.3 6.5
> At4g36650
Length=503
Score = 57.0 bits (136), Expect = 8e-09, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 54/96 (56%), Gaps = 9/96 (9%)
Query 6 LPRYCSRLQLSMHISDVAEHVAKRAA-QVFVSSHRPNSVAAAAIWLVVQLLSSSSAQQQM 64
+PR+C+ LQL+ ++A H+ + + F + P S++AAAI+L QL Q
Sbjct 233 MPRFCTLLQLNKSAQELATHIGEVVINKCFCTRRNPISISAAAIYLACQLEDKRKTQ--- 289
Query 65 LLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPN 100
+E+ +TG E TLR +YK++L+ + LLP+
Sbjct 290 -----AEICKITGLTEVTLRKVYKELLENWDDLLPS 320
> ECU06g1100i
Length=312
Score = 50.4 bits (119), Expect = 9e-07, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 53/97 (54%), Gaps = 8/97 (8%)
Query 4 HLLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQ 62
+++ R+CS L L++ I +A +AK A ++ ++ P+S+AAA I++V L Q
Sbjct 208 NIIARFCSDLNLNIKIQKIATEIAKAAHELGCLAGKSPDSIAAAVIYMVTNLFPEEKKIQ 267
Query 63 QMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
+ ++ VT E T+++ YK++L ++P
Sbjct 268 K-------DIQFVTNVTEVTIKNTYKELLTFKYDIIP 297
> 7297699
Length=315
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/95 (31%), Positives = 46/95 (48%), Gaps = 9/95 (9%)
Query 8 RYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQQMLL 66
R+C+ L L + A H+AK+A ++ V P SVAAAAI++ Q +Q+
Sbjct 216 RFCANLDLPNMVQRAATHIAKKAVEMDIVPGRSPISVAAAAIYMASQASEHKRSQK---- 271
Query 67 PKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
E+ + G + T+R YK M A L P +
Sbjct 272 ----EIGDIAGVADVTIRQSYKLMYPHAAKLFPED 302
> Hs4504193
Length=316
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 28/98 (28%), Positives = 47/98 (47%), Gaps = 9/98 (9%)
Query 5 LLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQQ 63
+ R+CS L L + A H+A++A ++ V P SVAAAAI++ Q + Q+
Sbjct 214 FMSRFCSNLCLPKQVQMAATHIARKAVELDLVPGRSPISVAAAAIYMASQASAEKRTQK- 272
Query 64 MLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
E+ + G + T+R Y+ + A L P +
Sbjct 273 -------EIGDIAGVADVTIRQSYRLIYPRAPDLFPTD 303
> CE27396
Length=306
Score = 47.4 bits (111), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 48/99 (48%), Gaps = 9/99 (9%)
Query 4 HLLPRYCSRLQLSMHISDVAEHVAKRAAQV-FVSSHRPNSVAAAAIWLVVQLLSSSSAQQ 62
+ R+C L L I A +AK A + V+ P S+AAAAI++ Q +SA++
Sbjct 203 DFMSRFCGNLSLPNSIQAAATRIAKCAVDMDLVAGRTPISIAAAAIYMASQ----ASAEK 258
Query 63 QMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPNE 101
+ E+ V GA E T+R YK + A L P +
Sbjct 259 R----SAKEIGDVAGAAEITVRQTYKLLYPKALELFPKD 293
> SPAC16E8.16
Length=340
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 29/89 (32%), Positives = 48/89 (53%), Gaps = 9/89 (10%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAA-QVFVSSHRPNSVAAAAIWLVVQLLSSSS 59
+A L+ R+C+RL L M + A +A+RA Q ++ P S+AA+ I+++ L+
Sbjct 231 SAEDLMVRFCNRLMLPMSVQSAAAELARRAGLQGTLAGRSPISIAASGIYMISALMGYPK 290
Query 60 AQQQMLLPKVSEVASVTGAGEHTLRSIYK 88
PK E++ VTG + T+R YK
Sbjct 291 T------PK--EISEVTGVSDSTIRIAYK 311
> At2g41630
Length=312
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 25/100 (25%), Positives = 47/100 (47%), Gaps = 9/100 (9%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSA 60
+A + R+CS L +S H A+ +++ + F P S+AA I+++ QL
Sbjct 208 HAGDFMRRFCSNLAMSNHAVKAAQEAVQKSEE-FDIRRSPISIAAVVIYIITQLSDDKKT 266
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPN 100
+ +++ TG E T+R+ YKD+ + P+
Sbjct 267 --------LKDISHATGVAEGTIRNSYKDLYPHLSKIAPS 298
> YPR086w
Length=345
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 50/94 (53%), Gaps = 9/94 (9%)
Query 6 LPRYCSRLQLSMHISDVAEHVAKRAAQVF-VSSHRPNSVAAAAIWLVVQLLSSSSAQQQM 64
+PR+CS L L M ++ AE+ AK+ ++ ++ P ++A +I+L + L Q +
Sbjct 239 IPRFCSHLGLPMQVTTSAEYTAKKCKEIKEIAGKSPITIAVVSIYLNILLF-----QIPI 293
Query 65 LLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLL 98
KV + VT E T++S YK + + + L+
Sbjct 294 TAAKVGQTLQVT---EGTIKSGYKILYEHRDKLV 324
> At3g10330
Length=312
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 51/99 (51%), Gaps = 9/99 (9%)
Query 1 NATHLLPRYCSRLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSA 60
+A + R+CS L ++ A+ +++ + F P S+AAA I+++ QL S
Sbjct 208 HAGDFMRRFCSNLGMTNQTVKAAQESVQKSEE-FDIRRSPISIAAAVIYIITQL----SD 262
Query 61 QQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLP 99
+++ L +++ TG E T+R+ YKD+ ++P
Sbjct 263 EKKPL----RDISVATGVAEGTIRNSYKDLYPHLSKIIP 297
> At3g29380
Length=336
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 30/101 (29%), Positives = 48/101 (47%), Gaps = 10/101 (9%)
Query 1 NATHLLPRYCSRLQLSM-HISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSS 59
N L+ R+CS+L +S I + E V K A+ F P SV AA I+++ +
Sbjct 219 NTGELVRRFCSKLDISQREIMAIPEAVEK--AENFDIRRNPKSVLAAIIFMISHI----- 271
Query 60 AQQQMLLPKVSEVASVTGAGEHTLRSIYKDMLDVAEHLLPN 100
Q + E+ V E+T+++ KDM A ++PN
Sbjct 272 --SQTNRKPIREIGIVAEVVENTIKNSVKDMYPYALKIIPN 310
> At1g71300
Length=701
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 42/85 (49%), Gaps = 4/85 (4%)
Query 12 RLQLSMHISDVAEHVAKRAAQVFVSSHRP--NSVAAAAIWLVVQLLSSSSAQQQMLLPKV 69
+LQL + S+ V R+ +F S P N A A+ +Q++ Q L+P +
Sbjct 296 KLQLDIATSNDLIGVDTRSTGLFSRSKEPLKNRCAVFALGERIQIIKE--IDQPALIPHI 353
Query 70 SEVASVTGAGEHTLRSIYKDMLDVA 94
+E +S+ E RS++K ++D A
Sbjct 354 AEASSLKYPYEVLFRSLHKLLMDTA 378
> Hs4758698
Length=1143
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 19 ISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSAQ 61
IS V+ +V+ QVF +S +P V +A+W +S ++ +
Sbjct 661 ISLVSVYVSSPTVQVFSASGKPVEVQVSAVWDTANTISETAYE 703
> Hs20472401
Length=1320
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 22 VAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSAQQQMLLPKVSEVASV 75
+AE + R +VF+ SH ++V+ + L +LLSS A++++++ +V + V
Sbjct 732 LAEVIKNRFHRVFLPSHSLDTVSPSDTLLCFELLSSELAKERVVVLEVQQRPQV 785
> At1g71270
Length=735
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 37/83 (44%), Gaps = 0/83 (0%)
Query 12 RLQLSMHISDVAEHVAKRAAQVFVSSHRPNSVAAAAIWLVVQLLSSSSAQQQMLLPKVSE 71
+LQL + + V R +F + P +A L ++ Q L+P ++E
Sbjct 330 KLQLDIATAYDLIGVETRTTGLFSRAREPLKNRSAVFALGDRIKIIKDIDQPALIPHIAE 389
Query 72 VASVTGAGEHTLRSIYKDMLDVA 94
+S+ E RS++K ++D A
Sbjct 390 ASSLKYPYEVLFRSLHKLLMDTA 412
Lambda K H
0.317 0.125 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40