bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1842_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
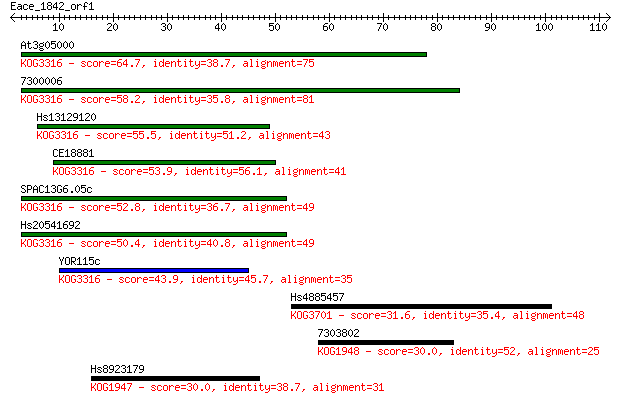
At3g05000 64.7 4e-11
7300006 58.2 4e-09
Hs13129120 55.5 2e-08
CE18881 53.9 7e-08
SPAC13G6.05c 52.8 2e-07
Hs20541692 50.4 8e-07
YOR115c 43.9 7e-05
Hs4885457 31.6 0.34
7303802 30.0 1.0
Hs8923179 30.0 1.1
> At3g05000
Length=173
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 41/75 (54%), Gaps = 0/75 (0%)
Query 3 RMPERRECVKFICKDLWHFLFQKQADRLQTNRRGGYVVHDANLPWLRRVHPSAAAPAATA 62
R + E +KFICKD W +F+KQ D L+TN RG +V+ D WL RV ++ T
Sbjct 58 RFSDHLEAIKFICKDFWSEVFKKQIDNLKTNHRGTFVLQDNKFRWLSRVSIDPSSENETQ 117
Query 63 TATTAAAAAAAVAAT 77
+T + AA A +
Sbjct 118 DPSTPGESKAAQAVS 132
> 7300006
Length=152
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 39/83 (46%), Gaps = 2/83 (2%)
Query 3 RMPERRECVKFICKDLWHFLFQKQADRLQTNRRGGYVVHDANLPWLRRVHPSAA--APAA 60
R + E +KFIC D W +++KQ D L+TN G YVV D +L R+ P A
Sbjct 51 RFKDELETMKFICTDFWMLIYKKQVDNLRTNNHGMYVVQDKAFRFLTRISPGTKQLEHAP 110
Query 61 TATATTAAAAAAAVAATGEPSEV 83
A T A++ G S V
Sbjct 111 KFVAFTCGLVRGALSNLGINSTV 133
> Hs13129120
Length=173
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 22/43 (51%), Positives = 30/43 (69%), Gaps = 0/43 (0%)
Query 6 ERRECVKFICKDLWHFLFQKQADRLQTNRRGGYVVHDANLPWL 48
E + +KF+CKDLW +FQKQ D L+TN +G YV+ D + P L
Sbjct 75 EELDVLKFLCKDLWVAVFQKQMDSLRTNHQGTYVLQDNSFPLL 117
> CE18881
Length=185
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 23/41 (56%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 9 ECVKFICKDLWHFLFQKQADRLQTNRRGGYVVHDANLPWLR 49
E VKFICKD W +F KQ D L+TN +G YVV D LR
Sbjct 90 EIVKFICKDFWSSVFGKQVDNLRTNHQGVYVVQDGRFTTLR 130
> SPAC13G6.05c
Length=245
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 32/49 (65%), Gaps = 0/49 (0%)
Query 3 RMPERRECVKFICKDLWHFLFQKQADRLQTNRRGGYVVHDANLPWLRRV 51
R+ E + ++F+C++LW +F+K D L+TNRRG +V+ D W ++
Sbjct 64 RITETTDVMRFLCRELWPIVFRKPLDNLKTNRRGIFVLTDTYFYWFTKM 112
> Hs20541692
Length=158
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 3 RMPERRECVKFICKDLWHFLFQKQADRLQTNRRGGYVVHDANLPWLRRV 51
R + + +KFICKD W +F+KQ D L+TN +G YV+ D L ++
Sbjct 57 RFKDELDIMKFICKDFWTTVFKKQIDNLRTNHQGIYVLQDNKFRLLTQM 105
> YOR115c
Length=268
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 10 CVKFICKDLWHFLFQKQADRLQTNRRGGYVVHDAN 44
+KFIC+D+W +F KQ D L+TN RG + + D +
Sbjct 156 IMKFICRDVWKQIFGKQIDNLKTNHRGTFYLLDYD 190
> Hs4885457
Length=552
Score = 31.6 bits (70), Expect = 0.34, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 53 PSAAAPAATATATTAAAAAAAVAATGEPSEVLQQQHEEELLR-AAATQP 100
P+ AP+ + +TA VA+T +P+ +L H E LL+ A+ QP
Sbjct 198 PALLAPSESNATSTANFPNIPVASTSQPASILGGSHSEGLLQIASGPQP 246
> 7303802
Length=1199
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 58 PAATATATTAAAAAAAVAATGEPSE 82
P ATAT T+A A+ ATG+P+E
Sbjct 493 PDATATCTSAEVTLQALDATGQPTE 517
> Hs8923179
Length=326
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 16 KDLWHFLFQKQADRLQTNRRGGYVVHDANLP 46
K +WH L + A RL + R GGY+ + P
Sbjct 57 KVMWHLLRRYMASRLHSLRMGGYLFSGSQAP 87
Lambda K H
0.323 0.129 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40