bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1851_orf2
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
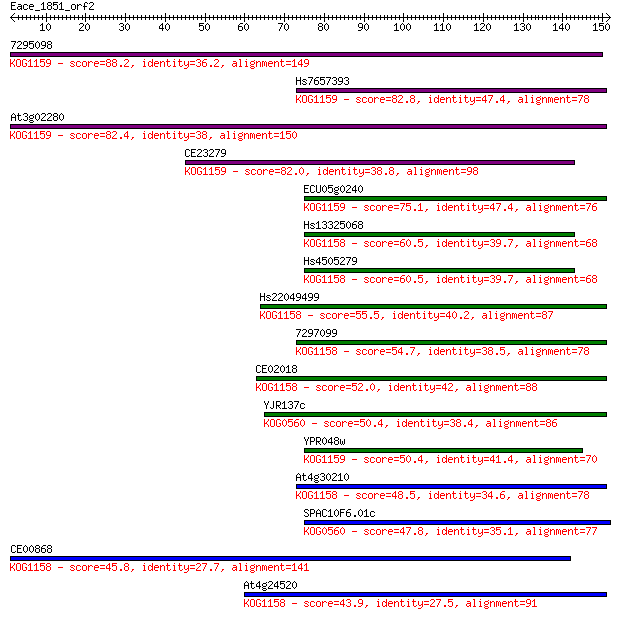
7295098 88.2 5e-18
Hs7657393 82.8 3e-16
At3g02280 82.4 3e-16
CE23279 82.0 4e-16
ECU05g0240 75.1 4e-14
Hs13325068 60.5 1e-09
Hs4505279 60.5 1e-09
Hs22049499 55.5 4e-08
7297099 54.7 6e-08
CE02018 52.0 5e-07
YJR137c 50.4 1e-06
YPR048w 50.4 1e-06
At4g30210 48.5 4e-06
SPAC10F6.01c 47.8 9e-06
CE00868 45.8 3e-05
At4g24520 43.9 1e-04
> 7295098
Length=582
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 54/149 (36%), Positives = 75/149 (50%), Gaps = 29/149 (19%)
Query 1 FATQTGNAQAAAADIWREAQRTLSLQQQPLLLPLAAAWPHLLQQQQQQQQQRQQQQQQQQ 60
+ +QTG AQ A IWRE+ + L Q +LP
Sbjct 7 YGSQTGTAQDVAEQIWRESHQ---LGFQGPVLPF-------------------------- 37
Query 61 QQDDLQRSPTKVVAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDR 120
+ D+ + + + +FV++TTG G PD+MK WR LL++ LP L +QFA GLGD
Sbjct 38 DEYDMTKLIEERLVVFVVATTGDGVEPDNMKLAWRFLLKRSLPAQSLQGMQFACLGLGDS 97
Query 121 RYVEFNFAAIKFYKRLQQLGAQPIVRLGL 149
Y +FN+AA K KRLQ LGA + +GL
Sbjct 98 SYPKFNYAAKKLSKRLQNLGANSVCPVGL 126
> Hs7657393
Length=597
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/78 (47%), Positives = 50/78 (64%), Gaps = 0/78 (0%)
Query 73 VAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVEFNFAAIKF 132
+ IFV +TTG GD PD+MK WR + ++ LP L + FAV GLGD Y +FNF A K
Sbjct 53 LVIFVCATTGQGDPPDNMKNFWRFIFRKNLPSTALCQMDFAVLGLGDSSYAKFNFVAKKL 112
Query 133 YKRLQQLGAQPIVRLGLG 150
++RL QLG ++ + LG
Sbjct 113 HRRLLQLGGSALLPVCLG 130
> At3g02280
Length=616
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 57/150 (38%), Positives = 72/150 (48%), Gaps = 35/150 (23%)
Query 1 FATQTGNAQAAAADIWREAQRTLSLQQQPLLLPLAAAWPHLLQQQQQQQQQRQQQQQQQQ 60
+A+QTGNA AA I REA+R LP +
Sbjct 12 YASQTGNALDAAERIGREAERRG--------LPASVV----------------------- 40
Query 61 QQDDLQRSPTKVVAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDR 120
D+ +FV+STTG GD PDS K WR LLQ+ L +L +++AVFGLGD
Sbjct 41 STDEFDTE----AVVFVVSTTGQGDSPDSFKAFWRFLLQRNLGNYWLQQVRYAVFGLGDS 96
Query 121 RYVEFNFAAIKFYKRLQQLGAQPIVRLGLG 150
Y ++NF A K KRL LGA I+ GLG
Sbjct 97 GYQKYNFVAKKLDKRLSDLGATTIIEKGLG 126
> CE23279
Length=585
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 38/98 (38%), Positives = 63/98 (64%), Gaps = 0/98 (0%)
Query 45 QQQQQQQRQQQQQQQQQQDDLQRSPTKVVAIFVISTTGYGDVPDSMKTLWRGLLQQQLPP 104
+++ QQ+ Q + + + D+ + P + V +FV+STTG G++P +M+ W+ LL++ L
Sbjct 22 RREAQQRHLQARVHELDEYDVSQLPMEKVVLFVVSTTGQGEMPPNMRKTWKLLLRKSLGA 81
Query 105 DYLSWLQFAVFGLGDRRYVEFNFAAIKFYKRLQQLGAQ 142
D+L + V GLGD Y ++NFA K Y+RL QLGA+
Sbjct 82 DFLKNVNIGVLGLGDSSYQKYNFAGKKLYRRLVQLGAK 119
> ECU05g0240
Length=510
Score = 75.1 bits (183), Expect = 4e-14, Method: Composition-based stats.
Identities = 36/76 (47%), Positives = 47/76 (61%), Gaps = 0/76 (0%)
Query 75 IFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVEFNFAAIKFYK 134
IFV ST G G P +M W L + LP LS L FAVFGLGD Y +FN+ + + +
Sbjct 69 IFVCSTHGDGAEPFNMTKFWSFLSRDDLPSTILSHLSFAVFGLGDSSYEKFNYCSKRLFN 128
Query 135 RLQQLGAQPIVRLGLG 150
RL+ LGA+P++R G G
Sbjct 129 RLRMLGARPVIRRGSG 144
> Hs13325068
Length=725
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 40/68 (58%), Gaps = 0/68 (0%)
Query 75 IFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVEFNFAAIKFYK 134
+ V+STTG GD PD+ + + + Q LP D+ + L++ + GLGD Y F K
Sbjct 81 VVVVSTTGTGDPPDTARKFVKEIQNQTLPVDFFAHLRYGLLGLGDSEYTYFCNGGKIIDK 140
Query 135 RLQQLGAQ 142
RLQ+LGA+
Sbjct 141 RLQELGAR 148
> Hs4505279
Length=698
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 40/68 (58%), Gaps = 0/68 (0%)
Query 75 IFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVEFNFAAIKFYK 134
+ V+STTG GD PD+ + + + Q LP D+ + L++ + GLGD Y F K
Sbjct 54 VVVVSTTGTGDPPDTARKFVKEIQNQTLPVDFFAHLRYGLLGLGDSEYTYFCNGGKIIDK 113
Query 135 RLQQLGAQ 142
RLQ+LGA+
Sbjct 114 RLQELGAR 121
> Hs22049499
Length=653
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 35/89 (39%), Positives = 49/89 (55%), Gaps = 5/89 (5%)
Query 64 DLQRSPT--KVVAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRR 121
DL P + +F ++T G GD D+ + + L + + LS ++FAVFGLG++
Sbjct 97 DLSSLPEIDNALVVFCMATYGEGDPTDNAQDFYDWLQETDVD---LSGVKFAVFGLGNKT 153
Query 122 YVEFNFAAIKFYKRLQQLGAQPIVRLGLG 150
Y FN KRL+QLGAQ I LGLG
Sbjct 154 YEHFNAMGKYVDKRLEQLGAQRIFELGLG 182
> 7297099
Length=679
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 30/78 (38%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 73 VAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVEFNFAAIKF 132
+A+F ++T G GD D+ + + + LS L +AVFGLG++ Y +N AI
Sbjct 136 LAVFCLATYGEGDPTDNAMEFYEWITSGDVD---LSGLNYAVFGLGNKTYEHYNKVAIYV 192
Query 133 YKRLQQLGAQPIVRLGLG 150
KRL++LGA + LGLG
Sbjct 193 DKRLEELGANRVFELGLG 210
> CE02018
Length=662
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 37/90 (41%), Positives = 50/90 (55%), Gaps = 5/90 (5%)
Query 63 DDLQRSPTKVVAIFV--ISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDR 120
+DL R A+ V I+T G GD D+ TL L LS ++FAVFGLG++
Sbjct 113 EDLNRLSEVEDALLVLCIATYGEGDPTDNAVTLVEYLNAGDCD---LSGVRFAVFGLGNK 169
Query 121 RYVEFNFAAIKFYKRLQQLGAQPIVRLGLG 150
Y FN I+ K+L++LGA+ I LGLG
Sbjct 170 TYEHFNEIGIQMDKQLEKLGAKRIFHLGLG 199
> YJR137c
Length=1442
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 33/93 (35%), Positives = 47/93 (50%), Gaps = 9/93 (9%)
Query 65 LQRSPTKVVAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVE 124
L+ P + +F+ ST G G+ P K+ W L+ D L+ L AVFGLGD Y
Sbjct 721 LEELPGEENVVFITSTAGQGEFPQDGKSFWEA-LKNDTDLD-LASLNVAVFGLGDSEYWP 778
Query 125 -------FNFAAIKFYKRLQQLGAQPIVRLGLG 150
FN + +KRL+ L A+ ++ LGLG
Sbjct 779 RKEDKHYFNKPSQDLFKRLELLSAKALIPLGLG 811
> YPR048w
Length=623
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 29/80 (36%), Positives = 47/80 (58%), Gaps = 10/80 (12%)
Query 75 IFVI-STTGYGDVPDSMK--------TLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVEF 125
+F+I STTG G++P ++ T W L ++ LP + L+ +Q A+ GLGD Y +F
Sbjct 55 LFIICSTTGQGELPRNVNALKGERPVTFWSFLKRKNLPSNLLNHIQTAMLGLGDSSYPKF 114
Query 126 NFAAIKFYKRL-QQLGAQPI 144
N+ K ++R+ QLGA +
Sbjct 115 NYGIRKLHQRIVTQLGANEL 134
> At4g30210
Length=711
Score = 48.5 bits (114), Expect = 4e-06, Method: Composition-based stats.
Identities = 27/78 (34%), Positives = 44/78 (56%), Gaps = 0/78 (0%)
Query 73 VAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRYVEFNFAAIKF 132
VA F ++T G G+ D+ ++ + ++L L++ VFGLG+R+Y FN A
Sbjct 160 VAFFFLATYGDGEPTDNAARFYKWFTEGNDRGEWLKNLKYGVFGLGNRQYEHFNKVAKVV 219
Query 133 YKRLQQLGAQPIVRLGLG 150
L + GAQ +V++GLG
Sbjct 220 DDILVEQGAQRLVQVGLG 237
> SPAC10F6.01c
Length=1473
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 42/84 (50%), Gaps = 10/84 (11%)
Query 75 IFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDRRY-------VEFNF 127
+ ++ST G G+ P + + W + L LS L++ VFG GD Y + +N
Sbjct 777 VVLVSTAGQGEFPQNGREFWEAIKGADLN---LSELKYGVFGFGDYEYWPRKEDKIYYNR 833
Query 128 AAIKFYKRLQQLGAQPIVRLGLGH 151
+ R +LGA P+V LGLG+
Sbjct 834 PGKQLDARFVELGAAPLVTLGLGN 857
> CE00868
Length=682
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 39/141 (27%), Positives = 62/141 (43%), Gaps = 28/141 (19%)
Query 1 FATQTGNAQAAAADIWREAQRTLSLQQQPLLLPLAAAWPHLLQQQQQQQQQRQQQQQQQQ 60
F +QTG A+ A SL+++ L+ L H L + +++ ++
Sbjct 8 FGSQTGQAETIAK----------SLKEKAELIGLTPRL-HALDENEKKFNLNEE------ 50
Query 61 QQDDLQRSPTKVVAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGDR 120
K+ AI V+S+TG GD PD+ R + + L +YL L + + GLGD
Sbjct 51 ----------KLCAI-VVSSTGDGDAPDNCARFVRRINRNSLENEYLKNLDYVLLGLGDS 99
Query 121 RYVEFNFAAIKFYKRLQQLGA 141
Y + K K+L LGA
Sbjct 100 NYSSYQTIPRKIDKQLTALGA 120
> At4g24520
Length=692
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 48/91 (52%), Gaps = 3/91 (3%)
Query 60 QQQDDLQRSPTKVVAIFVISTTGYGDVPDSMKTLWRGLLQQQLPPDYLSWLQFAVFGLGD 119
Q ++ L++ + +A F ++T G G+ D+ ++ ++ L L + VF LG+
Sbjct 130 QYEEKLKK---ETLAFFCVATYGDGEPTDNAARFYKWFTEENERDIKLQQLAYGVFALGN 186
Query 120 RRYVEFNFAAIKFYKRLQQLGAQPIVRLGLG 150
R+Y FN I + L + GA+ ++ +GLG
Sbjct 187 RQYEHFNKIGIVLDEELCKKGAKRLIEVGLG 217
Lambda K H
0.322 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40