bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1864_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
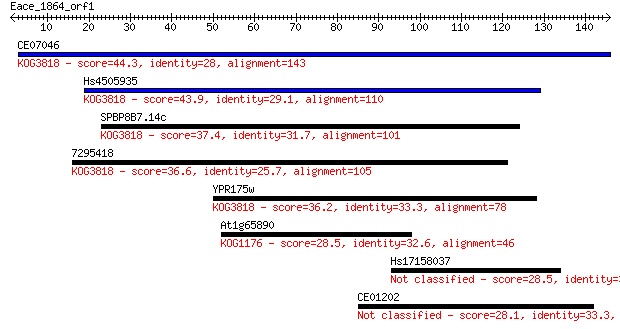
CE07046 44.3 9e-05
Hs4505935 43.9 1e-04
SPBP8B7.14c 37.4 0.010
7295418 36.6 0.016
YPR175w 36.2 0.023
At1g65890 28.5 4.4
Hs17158037 28.5 4.4
CE01202 28.1 5.3
> CE07046
Length=526
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 40/146 (27%), Positives = 57/146 (39%), Gaps = 12/146 (8%)
Query 3 RCCSSAALAAAKPSSTLWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVF 62
RC A AK T +S + + K ++ + + G + P VF
Sbjct 251 RCSDRLRSALAKQEDTSLVFLSDVFLDDKKVMKAVFKLLQGYKDQP--------PVAIVF 302
Query 63 LGNFEVNTKQRKDIK---AGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMP 119
GNF +Q I G + L N L + E FIFVPGP+DP F LPR
Sbjct 303 CGNFCSRPRQTDTIDLLDRGFRWLANQLTPLRKDYEKTQFIFVPGPDDP-FVDTVLPRPH 361
Query 120 IVSVYTQQFLNLMEQQFKGARDRIFF 145
+ S+ + ++ F RI F
Sbjct 362 LPSLLFKHISPIISCTFASNPCRIQF 387
> Hs4505935
Length=526
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 51/113 (45%), Gaps = 13/113 (11%)
Query 19 LWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNFE---VNTKQRKD 75
++ +S + + + L+ L +FAG P F+ GNF Q +
Sbjct 286 MFVFLSDVWLDQVEVLEKLRIMFAGYSPAP--------PTCFILCGNFSSAPYGKNQVQA 337
Query 76 IKAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQF 128
+K L+ L +++ +Y P F+FVPGP DP F LPR P+ T +F
Sbjct 338 LKDSLKTLADIICEY-PDIHQSRFVFVPGPEDPGFG-SILPRPPLAESITNEF 388
> SPBP8B7.14c
Length=594
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 44/105 (41%), Gaps = 11/105 (10%)
Query 23 ISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNFEV----NTKQRKDIKA 78
IS++ + LE IF + E S V P + G+F N+ K
Sbjct 335 ISEVHLDNHQTFYALEKIF-----QKYESSEAV-PFCIILCGSFMSSAFHNSGSSIQYKE 388
Query 79 GLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSV 123
G L L K+ + E FIFVPGPNDP + + MP S+
Sbjct 389 GFNKLAASLEKFPKICEKTKFIFVPGPNDP-WTTNGISLMPKHSI 432
> 7295418
Length=525
Score = 36.6 bits (83), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 42/105 (40%), Gaps = 9/105 (8%)
Query 16 SSTLWYIISQIQFGEPKDLQLLEAIFAGAISEAGEGSWGVLPAGFVFLGNFEVNTKQRKD 75
+ T +S ++ P ++ L +F G S P V +G F +T+ +
Sbjct 282 TETTIVFLSDVRLDLPVVMEKLRQLFVGYDS--------CPPQAIVLMGPFTASTRNHHE 333
Query 76 IKAGLQDLFNLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPI 120
++ L L L + L + I VP DP A LPR PI
Sbjct 334 LRHHLDALGGLAAGCEQLKKQTDLILVPSSEDPT-APNILPRAPI 377
> YPR175w
Length=692
Score = 36.2 bits (82), Expect = 0.023, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 37/84 (44%), Gaps = 9/84 (10%)
Query 50 EGSWGVLPAGFVFLGNFEVNTKQRKDIKAGLQDLFNLLLKYKPLAEHCYFIFVPGPND-- 107
+GS+ +P VF N K L LL ++ L E+ IF+PGPND
Sbjct 438 QGSFTSVP---VFASMSSRNISSSTQFKNNFDALATLLSRFDNLTENTTMIFIPGPNDLW 494
Query 108 ----PCFAREALPRMPIVSVYTQQ 127
A LP+ PI S +T++
Sbjct 495 GSMVSLGASGTLPQDPIPSAFTKK 518
> At1g65890
Length=578
Score = 28.5 bits (62), Expect = 4.4, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 21/46 (45%), Gaps = 9/46 (19%)
Query 52 SWGVLPAGFVFLGNFEVNTKQRKDIKAGLQDLFNLLLKYKPLAEHC 97
+WG P FV L E N + R+D L+ K + L E+C
Sbjct 480 TWGETPCAFVVLEKGETNNEDRED---------KLVTKERDLIEYC 516
> Hs17158037
Length=355
Score = 28.5 bits (62), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Query 93 LAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQFLNLME 133
L +HC+ + G + PC A P VS+ Q F+NL +
Sbjct 316 LCKHCHILSWKGTDHPCTANN--PESCSVSLSPQDFINLFK 354
> CE01202
Length=206
Score = 28.1 bits (61), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 12/57 (21%)
Query 85 NLLLKYKPLAEHCYFIFVPGPNDPCFAREALPRMPIVSVYTQQFLNLMEQQFKGARD 141
NLL+ Y+P A +FVP P PC R+ I + + +FK ARD
Sbjct 109 NLLVTYQPRALTHGHLFVPDPPTPC-------RLSITPCESGS-----DTEFKNARD 153
Lambda K H
0.326 0.141 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40