bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1900_orf1
Length=161
Score E
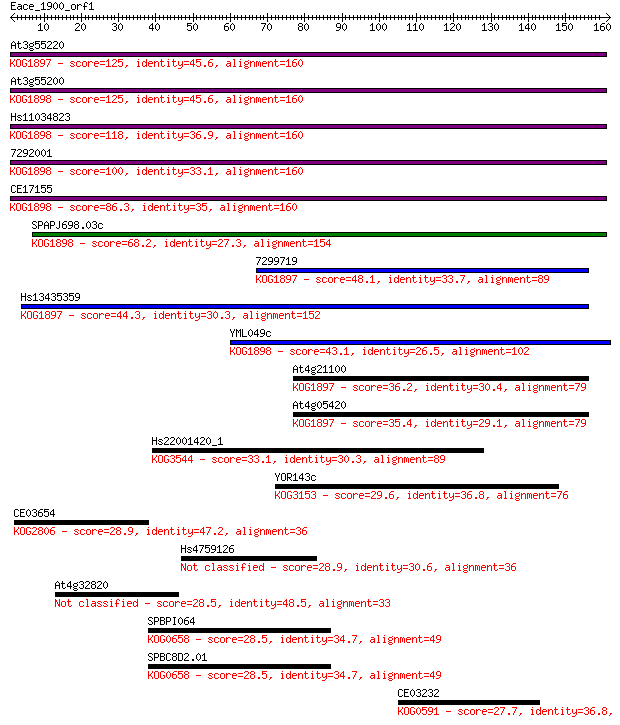
Sequences producing significant alignments: (Bits) Value
At3g55220 125 3e-29
At3g55200 125 3e-29
Hs11034823 118 5e-27
7292001 100 1e-21
CE17155 86.3 3e-17
SPAPJ698.03c 68.2 7e-12
7299719 48.1 8e-06
Hs13435359 44.3 1e-04
YML049c 43.1 3e-04
At4g21100 36.2 0.025
At4g05420 35.4 0.048
Hs22001420_1 33.1 0.24
YOR143c 29.6 2.7
CE03654 28.9 4.2
Hs4759126 28.9 4.6
At4g32820 28.5 5.3
SPBPI064 28.5 5.3
SPBC8D2.01 28.5 5.6
CE03232 27.7 8.9
> At3g55220
Length=1214
Score = 125 bits (314), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 73/165 (44%), Positives = 94/165 (56%), Gaps = 5/165 (3%)
Query 1 RAGEGKWGSCLRIVNPLLATTIEKLTYETDEAAISCCCCEF---EGLPLLIVGTVTAMTL 57
+A KW SC+R+++P ATT L + +EAA S C F E LL VGTV M
Sbjct 853 KAESEKWVSCIRVLDPKTATTTCLLELQDNEAAYSVCTVNFHDKEYGTLLAVGTVKGMQF 912
Query 58 RPKK-MAAASIKVYAY-DTNFRLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYALGK 115
PKK + A I +Y + + L LH T VE VP F+G LLA +G LRLY LGK
Sbjct 913 WPKKNLVAGFIHIYRFVEDGKSLELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGK 972
Query 116 KRLLKKCEYKNLPSTLVFLRVSGDRIFAGDMCESIHVLKYNAAAN 160
KRLL+KCE K P+T++ ++ DRI+ GD+ ES H KY N
Sbjct 973 KRLLRKCENKLFPNTIISIQTYRDRIYVGDIQESFHYCKYRRDEN 1017
> At3g55200
Length=1214
Score = 125 bits (314), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 73/165 (44%), Positives = 94/165 (56%), Gaps = 5/165 (3%)
Query 1 RAGEGKWGSCLRIVNPLLATTIEKLTYETDEAAISCCCCEF---EGLPLLIVGTVTAMTL 57
+A KW SC+R+++P ATT L + +EAA S C F E LL VGTV M
Sbjct 853 KAESEKWVSCIRVLDPKTATTTCLLELQDNEAAYSVCTVNFHDKEYGTLLAVGTVKGMQF 912
Query 58 RPKK-MAAASIKVYAY-DTNFRLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYALGK 115
PKK + A I +Y + + L LH T VE VP F+G LLA +G LRLY LGK
Sbjct 913 WPKKNLVAGFIHIYRFVEDGKSLELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGK 972
Query 116 KRLLKKCEYKNLPSTLVFLRVSGDRIFAGDMCESIHVLKYNAAAN 160
KRLL+KCE K P+T++ ++ DRI+ GD+ ES H KY N
Sbjct 973 KRLLRKCENKLFPNTIISIQTYRDRIYVGDIQESFHYCKYRRDEN 1017
> Hs11034823
Length=1217
Score = 118 bits (295), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 59/164 (35%), Positives = 92/164 (56%), Gaps = 4/164 (2%)
Query 1 RAGEGKWGSCLRIVNPLLATTIEKLTYETDEAAISCCCCEFEGLP---LLIVGTVTAMTL 57
+AG G+W S +R++NP+ T++ + E +EAA S C F ++VG + L
Sbjct 856 KAGNGQWASVIRVMNPIQGNTLDLVQLEQNEAAFSVAVCRFSNTGEDWYVLVGVAKDLIL 915
Query 58 RPKKMAAASIKVYAYDTNF-RLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYALGKK 116
P+ +A + Y N +L FLH TPVE+VP A F+G +L VG LR+Y LGKK
Sbjct 916 NPRSVAGGFVYTYKLVNNGEKLEFLHKTPVEEVPAAIAPFQGRVLIGVGKLLRVYDLGKK 975
Query 117 RLLKKCEYKNLPSTLVFLRVSGDRIFAGDMCESIHVLKYNAAAN 160
+LL+KCE K++ + + ++ G R+ D+ ES ++Y N
Sbjct 976 KLLRKCENKHIANYISGIQTIGHRVIVSDVQESFIWVRYKRNEN 1019
> 7292001
Length=1169
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 53/165 (32%), Positives = 88/165 (53%), Gaps = 5/165 (3%)
Query 1 RAGEGKWGSCLRIVNPLLATTIEKLTYETDEAAISCCCCEF----EGLPLLIVGTVTAMT 56
+AG G W S +R ++ + T+ + +EA +S +F +G L VG +
Sbjct 865 KAGLGLWASQIRCLDAMHGQTMFSVPLTQNEAIMSMAMLKFSIAADGRYYLAVGIAKDLQ 924
Query 57 LRPKKMAAASIKVYAYD-TNFRLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYALGK 115
L P+ I +Y D T L F+H T ++++P F+G LLA G LR+Y GK
Sbjct 925 LNPRISQGGCIDIYKIDPTCSSLEFMHRTDIDEIPGALCGFQGRLLAGCGRMLRIYDFGK 984
Query 116 KRLLKKCEYKNLPSTLVFLRVSGDRIFAGDMCESIHVLKYNAAAN 160
K++L+KCE K++P +V ++ G R++ D+ ES+ ++Y A N
Sbjct 985 KKMLRKCENKHIPYQIVNIQAMGHRVYVSDVQESVFFIRYRRAEN 1029
> CE17155
Length=1220
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 56/176 (31%), Positives = 83/176 (47%), Gaps = 19/176 (10%)
Query 1 RAGEGKWGSCLRIVNPLLATTIEKLTY---ETDEAAISCCCCEFEGLP---LLIVG---- 50
RA GKW S + +++ AT+ +KL+Y DE A +F P +++VG
Sbjct 850 RAARGKWASAISLIS---ATSGDKLSYFELPQDENAKCVALVQFSKHPNEAMVLVGCGVN 906
Query 51 -TVTAMTLRPKKMAAASIK--VYAYD---TNFRLTFLHSTPVEDVPQCFAAFRGMLLASV 104
+ + P + + VY + R FLH T FRGM L
Sbjct 907 EVLNVHDIDPNDTSIRPTRGCVYTFHLSANGDRFDFLHRTETPLPVGAIHDFRGMALVGF 966
Query 105 GNRLRLYALGKKRLLKKCEYKNLPSTLVFLRVSGDRIFAGDMCESIHVLKYNAAAN 160
G LR+Y +G+K+LL KCE KN P ++V ++ +G RI D ES+H L+Y N
Sbjct 967 GRFLRMYDIGQKKLLAKCENKNFPVSIVNIQSTGQRIIVSDSQESVHFLRYRKGDN 1022
> SPAPJ698.03c
Length=1206
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 42/157 (26%), Positives = 72/157 (45%), Gaps = 3/157 (1%)
Query 7 WGSCLRIVNPLLATTIEKLTYETDEAAISCCCCEFEGLP--LLIVGTVTAMTLRPKKMAA 64
W S + + + + I + +EAA S F+ L+ G+ T M L + +
Sbjct 853 WVSFISVFDMISKKIIHESPLGDNEAAFSMTAAFFKNRDEFFLVAGSATNMDLECRTCSH 912
Query 65 ASIKVYAY-DTNFRLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYALGKKRLLKKCE 123
+ +VY + D +L + T ++ +P F+G +LA VG LR+Y LG K++L+K E
Sbjct 913 GNFRVYRFHDEGKKLELISHTEIDGIPMALTPFQGRMLAGVGRFLRIYDLGNKKMLRKGE 972
Query 124 YKNLPSTLVFLRVSGDRIFAGDMCESIHVLKYNAAAN 160
+P + + V RI D S+ + Y N
Sbjct 973 LSAVPLFITHITVQASRIVVADSQYSVRFVVYKPEDN 1009
> 7299719
Length=1140
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 49/90 (54%), Gaps = 3/90 (3%)
Query 67 IKVYAYDTNFRLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYA-LGKKRLLKKCEYK 125
I ++ Y N +LT + T V+ F G +LA +G+ +RLY +K L +C +
Sbjct 849 IIIFHYHEN-KLTQVAETKVDGTCYALVEFNGKVLAGIGSFVRLYEWTNEKELRMECNIQ 907
Query 126 NLPSTLVFLRVSGDRIFAGDMCESIHVLKY 155
N+ + L FL+ GD I GD+ SI +L++
Sbjct 908 NMIAAL-FLKAKGDFILVGDLMRSITLLQH 936
> Hs13435359
Length=1140
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 46/165 (27%), Positives = 75/165 (45%), Gaps = 21/165 (12%)
Query 4 EGKWGSCLRIVNPLLATTIEKLTYET--------DEAAISCCCCEFEGLP--LLIVGT-- 51
E +G + + N L+ I++ T+E +E A+S C+ P IVGT
Sbjct 779 ETSFGEEVEVHNLLI---IDQHTFEVLHAHQFLQNEYALSLVSCKLGKDPNTYFIVGTAM 835
Query 52 VTAMTLRPKKMAAASIKVYAYDTNFRLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLY 111
V PK+ I V+ Y ++ +L + V+ F G LLAS+ + +RLY
Sbjct 836 VYPEEAEPKQ---GRIVVFQY-SDGKLQTVAEKEVKGAVYSMVEFNGKLLASINSTVRLY 891
Query 112 ALG-KKRLLKKCEYKNLPSTLVFLRVSGDRIFAGDMCESIHVLKY 155
+K + +C + N L +L+ GD I GD+ S+ +L Y
Sbjct 892 EWTTEKDVRTECNHYNNIMAL-YLKTKGDFILVGDLMRSVLLLAY 935
> YML049c
Length=1361
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 27/105 (25%), Positives = 52/105 (49%), Gaps = 3/105 (2%)
Query 60 KKMAAASIKVYAYDTNFRLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYALGKKRLL 119
K++ I V +F + LH T + F+ LL ++G+ + LY LGKK+LL
Sbjct 1001 KRLLTYKILVNKDKLSFDIELLHQTEIISPIHAMLKFKNFLLTAMGSTIVLYGLGKKQLL 1060
Query 120 KKCEYKNLPSTLVFLRV---SGDRIFAGDMCESIHVLKYNAAANL 161
++ + S + + + +R+ GD+ ES+ + ++ A N+
Sbjct 1061 RRSVTQTPVSITKIVSMHQWNYERLAVGDIHESVTLFIWDPAGNV 1105
> At4g21100
Length=1088
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 39/84 (46%), Gaps = 6/84 (7%)
Query 77 RLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYAL-----GKKRLLKKCEYKNLPSTL 131
RL + + AF G LLAS+ +++LY G + L +C + L
Sbjct 812 RLQLITEKETKGAVYSLNAFNGKLLASINQKIQLYKWMLRDDGTRELQSECGHHGHILAL 871
Query 132 VFLRVSGDRIFAGDMCESIHVLKY 155
+++ GD I GD+ +SI +L Y
Sbjct 872 -YVQTRGDFIAVGDLMKSISLLIY 894
> At4g05420
Length=1088
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 39/84 (46%), Gaps = 6/84 (7%)
Query 77 RLTFLHSTPVEDVPQCFAAFRGMLLASVGNRLRLYAL-----GKKRLLKKCEYKNLPSTL 131
RL + + AF G LLA++ +++LY G + L +C + L
Sbjct 812 RLQLIAEKETKGAVYSLNAFNGKLLAAINQKIQLYKWMLRDDGTRELQSECGHHGHILAL 871
Query 132 VFLRVSGDRIFAGDMCESIHVLKY 155
+++ GD I GD+ +SI +L Y
Sbjct 872 -YVQTRGDFIVVGDLMKSISLLLY 894
> Hs22001420_1
Length=324
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 36/92 (39%), Gaps = 3/92 (3%)
Query 39 CEFEGLPLLIVGTVTAMTLRPKKMAAASIKVYAYDTNFRLTFLHSTP---VEDVPQCFAA 95
C PL ++ + K + Y Y+TN R+T P VED F A
Sbjct 230 CPSRNAPLAVLSIPRVLQALTGKPEDNEVLKYPYETNIRVTLGPQPPCTEVEDAQFWFDA 289
Query 96 FRGMLLASVGNRLRLYALGKKRLLKKCEYKNL 127
R L VGN K+RL E++NL
Sbjct 290 SRKGLYLCVGNEWVSVLAAKERLDYVEEHQNL 321
> YOR143c
Length=319
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 39/84 (46%), Gaps = 16/84 (19%)
Query 72 YDTNFRLTFL---HSTPVEDVPQCFAAFRGMLLASVGNRLRLYALGKKRLLK-----KCE 123
Y T L FL + T +E PQ FR + + G L +G+ L+K K +
Sbjct 220 YMTPTDLIFLIKKNGTLIEYDPQ----FRNTCIGNCG----LLPIGEATLVKETRGLKWD 271
Query 124 YKNLPSTLVFLRVSGDRIFAGDMC 147
KN P+++V RVS F GD C
Sbjct 272 VKNWPTSVVTGRVSSSNRFVGDNC 295
> CE03654
Length=633
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 18/36 (50%), Gaps = 3/36 (8%)
Query 2 AGEGKWGSCLRIVNPLLATTIEKLTYETDEAAISCC 37
AGEGK G C R V PL E T D+A CC
Sbjct 501 AGEGKRGRCGRDVPPLEG---EAPTCNPDDANAHCC 533
> Hs4759126
Length=542
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 11/36 (30%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 47 LIVGTVTAMTLRPKKMAAASIKVYAYDTNFRLTFLH 82
+IVGT+ TLRP +M+ +K +++ + L
Sbjct 58 VIVGTILGFTLRPYRMSYREVKYFSFPGELLMRMLQ 93
> At4g32820
Length=1817
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 3/36 (8%)
Query 13 IVNPLLATTIEKLTYETDEAAI---SCCCCEFEGLP 45
+ NPL + EKL DEA SCC C FE +P
Sbjct 1081 LYNPLRFESWEKLGNIYDEAGSFIRSCCYCYFENVP 1116
> SPBPI064
Length=350
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 38 CCEFEGLPLLIVGTVTAMTLRPKKMAAASIKVYAYDTNFRLTFLHSTPV 86
C E +P + + T R K + SIK+YA+ L +LHST V
Sbjct 67 CLLLEYMPETVFDDMRWYTRRRKSIPNLSIKLYAFQLFRALAYLHSTGV 115
> SPBC8D2.01
Length=381
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 38 CCEFEGLPLLIVGTVTAMTLRPKKMAAASIKVYAYDTNFRLTFLHSTPV 86
C E +P + + T R K + SIK+YA+ L +LHST V
Sbjct 98 CLLLEYMPETVFDDMRWYTRRRKSIPNLSIKLYAFQLFRALAYLHSTGV 146
> CE03232
Length=294
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query 105 GNRLRLYALGKKRLLKKCEYKNLPSTLVFLRVSGDRIF 142
G+++ LY+L KK ++ CEY LP+ + +VS + F
Sbjct 227 GDKMNLYSLCKK--IENCEYPPLPADIYSTQVSANLCF 262
Lambda K H
0.325 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2208717646
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40