bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1929_orf2
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
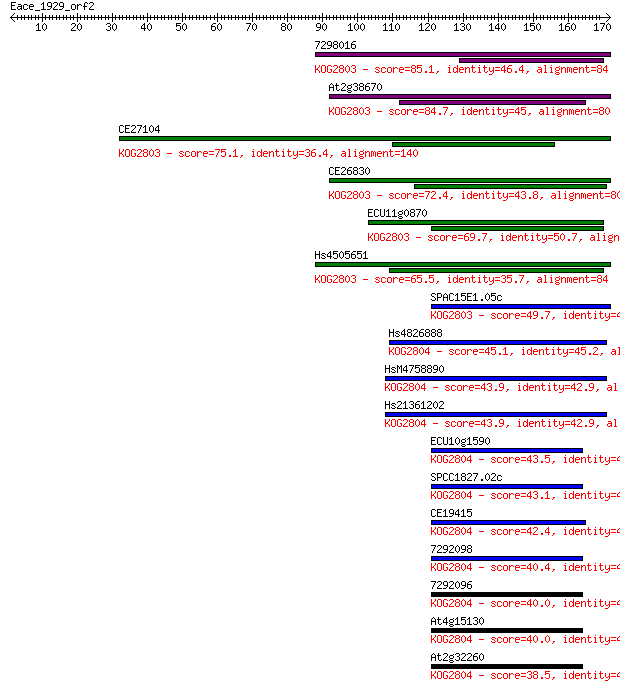
7298016 85.1 6e-17
At2g38670 84.7 8e-17
CE27104 75.1 7e-14
CE26830 72.4 4e-13
ECU11g0870 69.7 3e-12
Hs4505651 65.5 5e-11
SPAC15E1.05c 49.7 3e-06
Hs4826888 45.1 8e-05
HsM4758890 43.9 1e-04
Hs21361202 43.9 2e-04
ECU10g1590 43.5 2e-04
SPCC1827.02c 43.1 2e-04
CE19415 42.4 4e-04
7292098 40.4 0.002
7292096 40.0 0.002
At4g15130 40.0 0.002
At2g32260 38.5 0.007
> 7298016
Length=363
Score = 85.1 bits (209), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 39/85 (45%), Positives = 58/85 (68%), Gaps = 2/85 (2%)
Query 88 ASQPSRSTNLFL-SSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGD 146
A P + FL ++++++QF K P GD+IVYV G+FD+ HVGH+ L+ AK++GD
Sbjct 164 AKSPWTGCSQFLPTTQKIIQF-SDGKSPNPGDKIVYVAGAFDLFHVGHLDFLEKAKKLGD 222
Query 147 YLIVGIHDDATVAAVKGPGFPVLNL 171
YLIVG+H D V + KG +P++NL
Sbjct 223 YLIVGLHTDPVVNSYKGSNYPIMNL 247
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Query 129 VLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVL 169
++H GH L+ AK +GD +IVGIH D + KGP PV
Sbjct 1 MVHFGHANSLRQAKALGDKVIVGIHTDEEITKHKGP--PVF 39
> At2g38670
Length=421
Score = 84.7 bits (208), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 58/80 (72%), Gaps = 1/80 (1%)
Query 92 SRSTNLFLSSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVG 151
+R ++ +S+R++QF K P RI+Y+DG+FD+ H GHV IL+ A+++GD+L+VG
Sbjct 229 TRVSHFLPTSRRIVQF-SNGKGPGPDARIIYIDGAFDLFHAGHVEILRRARELGDFLLVG 287
Query 152 IHDDATVAAVKGPGFPVLNL 171
IH+D TV+A +G P++NL
Sbjct 288 IHNDQTVSAKRGAHRPIMNL 307
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 34/53 (64%), Gaps = 4/53 (7%)
Query 112 KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGP 164
+KP + VY+DG FD++H GH L+ A+ +GD L+VG+ D + A KGP
Sbjct 52 RKPVR----VYMDGCFDMMHYGHCNALRQARALGDQLVVGVVSDEEIIANKGP 100
> CE27104
Length=302
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 51/142 (35%), Positives = 68/142 (47%), Gaps = 9/142 (6%)
Query 32 TEETETAASEAASAEDTAAAAAAAPATGASAASTSSSASAAAAGGQTAEDPTAAAHASQP 91
T+E A D A T T S +A A DP + ++P
Sbjct 67 TDEERYRLISAIRWVDEEIAGRLMLVTKKHHMETDSILDFKSAILPFALDPLS----NEP 122
Query 92 SRSTNLFLSSKRLLQF--IGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLI 149
S +LF K+ F + +KPK D++VYV G+FD+ H GH+ L+ AK +GDYLI
Sbjct 123 VISVSLF---KQNYTFAPVVIGRKPKVTDKVVYVSGAFDLFHAGHLSFLEAAKDLGDYLI 179
Query 150 VGIHDDATVAAVKGPGFPVLNL 171
VGI D V KG FPV+NL
Sbjct 180 VGIVGDDDVNEEKGTIFPVMNL 201
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/46 (54%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 110 TPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDD 155
TP KK R VY DG FD +H + R+L AKQ G LIVGIH D
Sbjct 10 TPDGEKKKAR-VYTDGCFDFVHFANARLLWPAKQYGKKLIVGIHSD 54
> CE26830
Length=370
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 52/81 (64%), Gaps = 3/81 (3%)
Query 92 SRSTNLFLSSKRLLQFI-GTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIV 150
+R + ++ +L+F G P KP D++VYV GSFD+ H+GH+ L+ AK+ GDYLIV
Sbjct 178 TRVSRFIPTTTTILEFAEGRPPKPT--DKVVYVTGSFDLFHIGHLAFLEKAKEFGDYLIV 235
Query 151 GIHDDATVAAVKGPGFPVLNL 171
GI D TV KG P++++
Sbjct 236 GILSDQTVNQYKGSNHPIMSI 256
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 36/55 (65%), Gaps = 3/55 (5%)
Query 116 KGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVLN 170
KG+R V+ DG +D++H GH L+ AKQ G LIVG+H+D + KGP PV N
Sbjct 11 KGNR-VWADGCYDMVHFGHANQLRQAKQFGQKLIVGVHNDEEIRLHKGP--PVFN 62
> ECU11g0870
Length=322
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 34/68 (50%), Positives = 47/68 (69%), Gaps = 3/68 (4%)
Query 103 RLLQ-FIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAV 161
RLL F+ + + K+G ++V++DG+FD+ H GHV L+IA+ MGDYLIVGIHDD T
Sbjct 162 RLLSLFMSSMGREKRG-KVVFMDGNFDLFHAGHVASLRIARGMGDYLIVGIHDDETTKEY 220
Query 162 KGPGFPVL 169
+PVL
Sbjct 221 T-RSYPVL 227
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 30/49 (61%), Gaps = 2/49 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVL 169
V+ DG FD+ H GH L+ +K +GDYLI G+H ++ K G PV+
Sbjct 9 VWADGCFDMFHYGHANALRQSKALGDYLIAGVHSSLSINQEK--GLPVM 55
> Hs4505651
Length=389
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 54/87 (62%), Gaps = 4/87 (4%)
Query 88 ASQPSRSTNLFL-SSKRLLQFIGTPKKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQMGD 146
P + FL +S++++QF + K+P+ G+ ++YV G+FD+ H+GHV L+ ++ +
Sbjct 183 GRNPWTGVSQFLQTSQKIIQF-ASGKEPQPGETVIYVAGAFDLFHIGHVDFLEKVHRLAE 241
Query 147 --YLIVGIHDDATVAAVKGPGFPVLNL 171
Y+I G+H D V KG +P++NL
Sbjct 242 RPYIIAGLHFDQEVNHYKGKNYPIMNL 268
Score = 58.5 bits (140), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 29/64 (45%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 109 GTPKKPKKGDRI---VYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPG 165
G ++P G R V+ DG +D++H GH L+ A+ MGDYLIVG+H D +A KGP
Sbjct 10 GGAEQPGPGGRRAVRVWCDGCYDMVHYGHSNQLRQARAMGDYLIVGVHTDEEIAKHKGP- 68
Query 166 FPVL 169
PV
Sbjct 69 -PVF 71
> SPAC15E1.05c
Length=365
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 2/51 (3%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGDYLIVGIHDDATVAAVKGPGFPVLNL 171
+++DG D H GH + AKQ+G+ L++GIH D + KGP PV+ L
Sbjct 11 LWLDGCMDFFHYGHSNAILQAKQLGETLVIGIHSDEEITLNKGP--PVMTL 59
> Hs4826888
Length=367
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Query 109 GTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGPG 165
GTP ++P + VY DG FD+ H GH R L AK + YLIVG+ D K G
Sbjct 70 GTPCERPVR----VYADGIFDLFHSGHARALMQAKNLFPNTYLIVGVCSDELTHNFK--G 123
Query 166 FPVLN 170
F V+N
Sbjct 124 FTVMN 128
> HsM4758890
Length=330
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 35/66 (53%), Gaps = 9/66 (13%)
Query 108 IGTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGP 164
+GTP +P + VY DG FD+ H GH R L AK + YL+VG+ D K
Sbjct 69 LGTPADRPVR----VYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFK-- 122
Query 165 GFPVLN 170
GF V+N
Sbjct 123 GFTVMN 128
> Hs21361202
Length=369
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 35/66 (53%), Gaps = 9/66 (13%)
Query 108 IGTP-KKPKKGDRIVYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKGP 164
+GTP +P + VY DG FD+ H GH R L AK + YL+VG+ D K
Sbjct 69 LGTPADRPVR----VYADGIFDLFHSGHARALMQAKTLFPNSYLLVGVCSDDLTHKFK-- 122
Query 165 GFPVLN 170
GF V+N
Sbjct 123 GFTVMN 128
> ECU10g1590
Length=278
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
+Y DG +D+ H GH R LK AK + YL+VG+ DD +KG
Sbjct 45 IYCDGVYDLFHYGHARSLKQAKNLFPNVYLLVGVTDDDITVRLKG 89
> SPCC1827.02c
Length=354
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 30/45 (66%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
VY DG FD+ H+GH+R L+ AK++ +LIVG+ +D +KG
Sbjct 96 VYADGVFDLFHIGHMRQLEQAKKVFPNVHLIVGLPNDQLTHRLKG 140
> CE19415
Length=183
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQMGD--YLIVGIHDDATVAAVKGP 164
VY DG +D+ H GH + L KQM YLIVG+ D KGP
Sbjct 44 VYADGIYDMFHYGHAKQLLQIKQMFPMVYLIVGVCSDENTLKFKGP 89
> 7292098
Length=381
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
VY DG +D+ H GH R L AK + YLIVG+ +D +KG
Sbjct 81 VYADGIYDLFHQGHARQLMQAKNVFPNVYLIVGVCNDELTLRMKG 125
> 7292096
Length=347
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
VY DG +D+ H GH R L AK + YLIVG+ +D +KG
Sbjct 29 VYADGIYDLFHQGHARQLMQAKNIFPNVYLIVGVCNDELTHRMKG 73
> At4g15130
Length=298
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 25/45 (55%), Gaps = 2/45 (4%)
Query 121 VYVDGSFDVLHVGHVRILKIAKQM--GDYLIVGIHDDATVAAVKG 163
VY DG FD+ H GH R ++ AK+ YL+VG +D KG
Sbjct 23 VYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKG 67
> At2g32260
Length=332
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query 121 VYVDGSFDVLHVGHVRIL---KIAKQMGDYLIVGIHDDATVAAVKG 163
VY DG +D+ H GH R L K+A YL+VG +D T KG
Sbjct 37 VYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKG 82
Lambda K H
0.310 0.121 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2526689620
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40