bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_1930_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
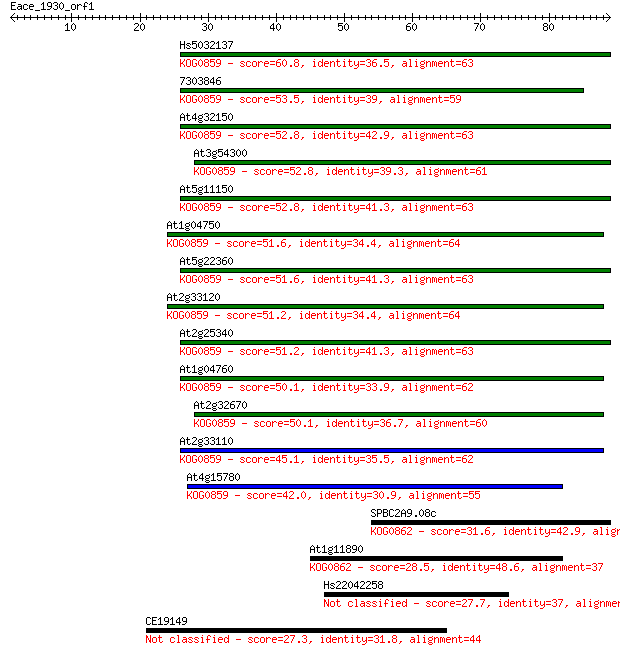
Hs5032137 60.8 5e-10
7303846 53.5 9e-08
At4g32150 52.8 2e-07
At3g54300 52.8 2e-07
At5g11150 52.8 2e-07
At1g04750 51.6 3e-07
At5g22360 51.6 3e-07
At2g33120 51.2 5e-07
At2g25340 51.2 5e-07
At1g04760 50.1 1e-06
At2g32670 50.1 1e-06
At2g33110 45.1 3e-05
At4g15780 42.0 3e-04
SPBC2A9.08c 31.6 0.36
At1g11890 28.5 3.3
Hs22042258 27.7 4.7
CE19149 27.3 7.1
> Hs5032137
Length=220
Score = 60.8 bits (146), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCEGL 85
M +++A++ARG +L +HA GNF VT +L ++P++ +++Y +G Y+FHYI + +
Sbjct 1 MAILFAVVARGTTILAKHAWCGGNFLEVTEQILAKIPSENNKLTYSHGNYLFHYICQDRI 60
Query 86 TFL 88
+L
Sbjct 61 VYL 63
> 7303846
Length=218
Score = 53.5 bits (127), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCEG 84
MP++Y++I+RG VL + A+ GNF VT ++ R+ +M+Y +G Y+ HY CE
Sbjct 1 MPILYSVISRGTTVLAKFAECVGNFAEVTEHIIGRIGVHNHKMTYTHGDYLIHY-TCEN 58
> At4g32150
Length=219
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 38/64 (59%), Gaps = 1/64 (1%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQ-RMSYIYGRYVFHYIVCEG 84
M ++YAL+ARG VVL E + N T+ + +L ++P D +SY RYVFH +G
Sbjct 1 MAILYALVARGTVVLSEFTATSTNASTIAKQILEKVPGDNDSNVSYSQDRYVFHVKRTDG 60
Query 85 LTFL 88
LT L
Sbjct 61 LTVL 64
> At3g54300
Length=240
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 36/61 (59%), Gaps = 0/61 (0%)
Query 28 LIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCEGLTF 87
LIY+ +A+G VVL EH GNF T+ L +LPT+ + +Y + F+++V G F
Sbjct 6 LIYSFVAKGTVVLAEHTPYSGNFSTIAVQCLQKLPTNSSKYTYSCDGHTFNFLVDNGFVF 65
Query 88 L 88
L
Sbjct 66 L 66
> At5g11150
Length=221
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTD--KQRMSYIYGRYVFHYIVCE 83
M +I+AL+ARG VVL E + + N ++++ +L +LP + MSY RY+FH +
Sbjct 1 MAIIFALVARGTVVLSEFSATSTNASSISKQILEKLPGNDSDSHMSYSQDRYIFHVKRTD 60
Query 84 GLTFL 88
GLT L
Sbjct 61 GLTVL 65
> At1g04750
Length=219
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 24 PTMPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCE 83
LIY+ +ARG V+L E D GNF ++ L +LP+ + +Y + F+Y+V +
Sbjct 2 AQQSLIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKFTYNCDGHTFNYLVED 61
Query 84 GLTF 87
G T+
Sbjct 62 GFTY 65
> At5g22360
Length=221
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTD--KQRMSYIYGRYVFHYIVCE 83
M ++YA++ARG VVL E + GN V R +L +L + +R+ + RY+FH + +
Sbjct 1 MAIVYAVVARGTVVLAEFSAVTGNTGAVVRRILEKLSPEISDERLCFSQDRYIFHILRSD 60
Query 84 GLTFL 88
GLTFL
Sbjct 61 GLTFL 65
> At2g33120
Length=221
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 24 PTMPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCE 83
LIY+ +ARG V+L E D GNF ++ L +LP+ + +Y + F+Y+V
Sbjct 2 AQQSLIYSFVARGTVILVEFTDFKGNFTSIAAQCLQKLPSSNNKFTYNCDGHTFNYLVEN 61
Query 84 GLTF 87
G T+
Sbjct 62 GFTY 65
> At2g25340
Length=219
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTD-KQRMSYIYGRYVFHYIVCEG 84
M ++YAL+ARG VVL E + + N T+ + +L ++P + +SY RYVFH +G
Sbjct 1 MSILYALVARGTVVLAELSTTSTNASTIAKQILEKIPGNGDSHVSYSQDRYVFHVKRTDG 60
Query 85 LTFL 88
LT L
Sbjct 61 LTVL 64
> At1g04760
Length=220
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 35/62 (56%), Gaps = 0/62 (0%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCEGL 85
LIY+ +ARG V+L E+ + GNF +V L +LP+ + +Y + F+Y+ G
Sbjct 4 QSLIYSFVARGTVILAEYTEFKGNFTSVAAQCLQKLPSSNNKFTYNCDGHTFNYLADNGF 63
Query 86 TF 87
T+
Sbjct 64 TY 65
> At2g32670
Length=220
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 35/60 (58%), Gaps = 0/60 (0%)
Query 28 LIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCEGLTF 87
LIY+ +ARG V+L E+ + GNF V L +LP+ + +Y + F+Y+V G T+
Sbjct 6 LIYSFVARGTVILVEYTEFKGNFTAVAAQCLQKLPSSNNKFTYNCDGHTFNYLVENGFTY 65
> At2g33110
Length=212
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 26 MPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQRMSYIYGRYVFHYIVCEGL 85
L Y+ IARG V+L E D GNF +V L LP+ + +Y + F+ +V G
Sbjct 4 QSLFYSFIARGTVILVEFTDFKGNFTSVAAQYLENLPSSNNKFTYNCDGHTFNDLVENGF 63
Query 86 TF 87
T+
Sbjct 64 TY 65
> At4g15780
Length=159
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 33/56 (58%), Gaps = 1/56 (1%)
Query 27 PLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTDKQ-RMSYIYGRYVFHYIV 81
IY+ +ARG ++L E+ + GNFP++ L +LP+ + +Y + F+++V
Sbjct 5 SFIYSFVARGTMILAEYTEFTGNFPSIAAQCLQKLPSSSNSKFTYNCDHHTFNFLV 60
> SPBC2A9.08c
Length=209
Score = 31.6 bits (70), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Query 54 TRLLLCRL-PTDKQRMSYIYGRYVFHYIVCEGLTFL 88
+L+L RL PT ++R S G Y FHY++ G+ +L
Sbjct 29 AKLILKRLSPTSEKRASIESGDYTFHYLIDNGICYL 64
> At1g11890
Length=218
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 19/39 (48%), Gaps = 2/39 (5%)
Query 45 DSDGNFPTVTRLL--LCRLPTDKQRMSYIYGRYVFHYIV 81
DSD V L L R D RMS G YVFHYI+
Sbjct 28 DSDMYKQQVKALFKNLSRGQNDASRMSVETGPYVFHYII 66
> Hs22042258
Length=546
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 47 DGNFPTVTRLLLCRLPTDKQRMSYIYG 73
DGN P+ + +PT Q +SY++G
Sbjct 112 DGNIPSQANVHCSSVPTGDQSLSYVHG 138
> CE19149
Length=424
Score = 27.3 bits (59), Expect = 7.1, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 20/44 (45%), Gaps = 0/44 (0%)
Query 21 VKAPTMPLIYALIARGNVVLCEHADSDGNFPTVTRLLLCRLPTD 64
V A T+P+ +A + E GN+P + LL C P D
Sbjct 218 VLAETIPVALPTLAMSMETMIEGRPLAGNYPVTSELLQCTPPLD 261
Lambda K H
0.330 0.147 0.465
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184307974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40