bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2044_orf2
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
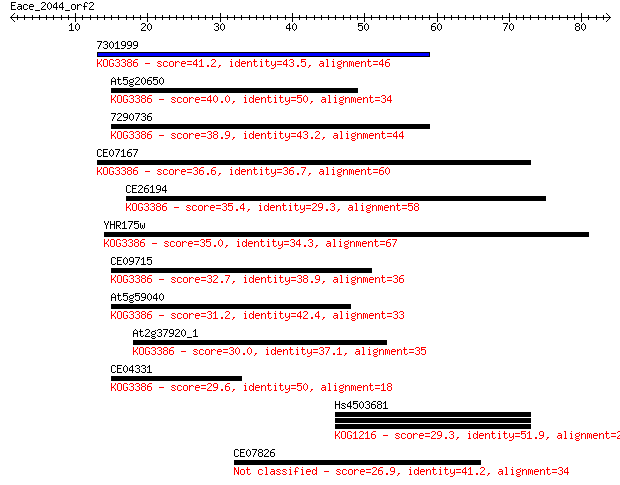
7301999 41.2 5e-04
At5g20650 40.0 0.001
7290736 38.9 0.003
CE07167 36.6 0.011
CE26194 35.4 0.029
YHR175w 35.0 0.030
CE09715 32.7 0.17
At5g59040 31.2 0.52
At2g37920_1 30.0 0.97
CE04331 29.6 1.6
Hs4503681 29.3 2.0
CE07826 26.9 9.1
> 7301999
Length=270
Score = 41.2 bits (95), Expect = 5e-04, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 13 MEMQMTFYWGYDATILFPWWETSTALEFYLSCLCIFALCLGSAKLK 58
M M M F+ G TILF +W T +A+ LSCL IF + + LK
Sbjct 61 MSMAMFFHTGDSETILFKFWRTESAMALTLSCLLIFMVAVLYEALK 106
> At5g20650
Length=146
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 15 MQMTFYWGYDATILFPWWETSTALEFYLSCLCIF 48
M MTFYWG ATILF +W+T + L + L+ + F
Sbjct 2 MHMTFYWGIKATILFDFWKTDSWLSYILTLIACF 35
> 7290736
Length=220
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 15 MQMTFYWGYDATILFPWWETSTALEFYLSCLCIFALCLGSAKLK 58
M M F++GY+ TILF WW T S + IF L L LK
Sbjct 58 MPMAFHFGYNETILFSWWHIETVAGLIGSMIAIFLLALMYEGLK 101
> CE07167
Length=162
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Query 13 MEMQMTFYWGYDATILFPWWETSTALEFYLSCLCIFALCLGSAKLK------AVCRDLQR 66
M+M MT ++G ILF WW+T + +S L F LC+ +K AV + +R
Sbjct 1 MDMDMTLHFGEREKILFSWWKTGSLSGMAVSMLITFLLCILYEAIKSFRYFLAVWNNQKR 60
Query 67 GGAHVE 72
H E
Sbjct 61 QQRHAE 66
> CE26194
Length=130
Score = 35.4 bits (80), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 31/58 (53%), Gaps = 1/58 (1%)
Query 17 MTFYWGYDATILFPWWETSTALEFYLSCLCIFALCLGSAKLKAVCRDLQRGGAHVEEP 74
M+F++G + TILF +W+T TA+ ++C L L+ RD ++ + +P
Sbjct 1 MSFHFGTEETILFDFWKTETAVGIAVACFITVLLAFLMETLRFF-RDYRKAQTQLHQP 57
> YHR175w
Length=189
Score = 35.0 bits (79), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 29/68 (42%), Gaps = 2/68 (2%)
Query 14 EMQMTFYWGYDAT-ILFPWWETSTALEFYLSCLCIFALCLGSAKLKAVCRDLQRGGAHVE 72
M M F W Y T ++F WW T LSCL IF L LK C ++ V
Sbjct 58 SMNMLFSWSYKNTCVVFEWWHIKTLPGLILSCLAIFGLAYLYEYLK-YCVHKRQLSQRVL 116
Query 73 EPARPVVR 80
P R + +
Sbjct 117 LPNRSLTK 124
> CE09715
Length=252
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 15 MQMTFYWGYDATILFPWWETSTALEFYLSCLCIFAL 50
M+M F+ G++ ILF +W T + LSC IF +
Sbjct 76 MKMWFHGGFEEVILFDFWRTDSLFGMLLSCAAIFIM 111
> At5g59040
Length=151
Score = 31.2 bits (69), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query 15 MQMTFYWGYDATILFPWWETSTALEFYLSCLCI 47
M MTF+WG +LF W T+L+ Y CL +
Sbjct 30 MHMTFFWGKTTEVLFDGWP-GTSLKMYWVCLAV 61
> At2g37920_1
Length=185
Score = 30.0 bits (66), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 18 TFYWGYDATILFPWWETSTALEFYLSCLCIFALCL 52
TFYWGY+ +LF W S + L+ + +F L
Sbjct 33 TFYWGYNCQVLFSGWPGSDRGMYALALIFVFFLAF 67
> CE04331
Length=178
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 9/18 (50%), Positives = 14/18 (77%), Gaps = 0/18 (0%)
Query 15 MQMTFYWGYDATILFPWW 32
M+M F+WG+D +LF +W
Sbjct 118 MKMWFHWGFDEVVLFDFW 135
> Hs4503681
Length=5405
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 17/34 (50%), Gaps = 7/34 (20%)
Query 46 CIFALCLGSAKLKAVCRDL-------QRGGAHVE 72
CIF LCLG L +C ++ Q G HVE
Sbjct 1487 CIFDLCLGGGNLSILCSNIHAYVSACQAAGGHVE 1520
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 17/34 (50%), Gaps = 7/34 (20%)
Query 46 CIFALCLGSAKLKAVCRDL-------QRGGAHVE 72
CIF LCLG L +C ++ Q G HVE
Sbjct 2688 CIFDLCLGGGNLSILCSNIHAYVSACQAAGGHVE 2721
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 17/34 (50%), Gaps = 7/34 (20%)
Query 46 CIFALCLGSAKLKAVCRDL-------QRGGAHVE 72
CIF LCLG L +C ++ Q G HVE
Sbjct 3889 CIFDLCLGGGNLSILCSNIHAYVSACQAAGGHVE 3922
> CE07826
Length=922
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 18/36 (50%), Gaps = 2/36 (5%)
Query 32 WETSTALEFYLSCLCIFALC--LGSAKLKAVCRDLQ 65
W S ++FYL CIF + L S LK +C Q
Sbjct 138 WSLSLEMQFYLFAPCIFFMLQFLNSKYLKVLCATRQ 173
Lambda K H
0.328 0.139 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200681374
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40