bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2054_orf1
Length=97
Score E
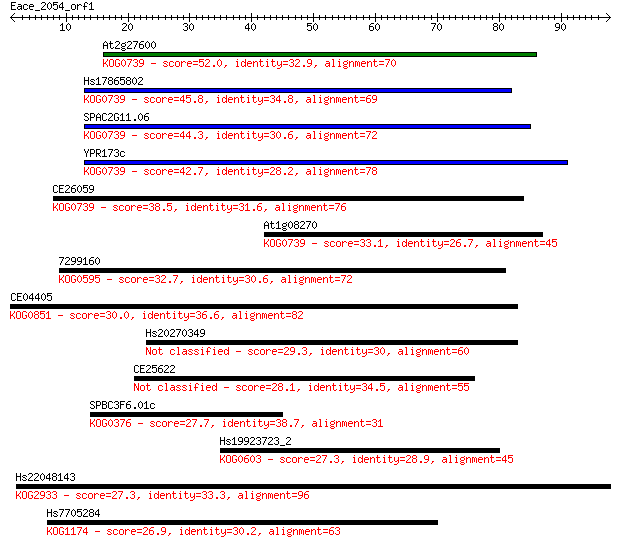
Sequences producing significant alignments: (Bits) Value
At2g27600 52.0 3e-07
Hs17865802 45.8 2e-05
SPAC2G11.06 44.3 6e-05
YPR173c 42.7 2e-04
CE26059 38.5 0.003
At1g08270 33.1 0.12
7299160 32.7 0.17
CE04405 30.0 1.1
Hs20270349 29.3 1.6
CE25622 28.1 4.2
SPBC3F6.01c 27.7 5.4
Hs19923723_2 27.3 6.8
Hs22048143 27.3 7.7
Hs7705284 26.9 8.4
> At2g27600
Length=435
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 45/70 (64%), Gaps = 0/70 (0%)
Query 16 AIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQ 75
AI +A +D+AG+ +AF LY ALE ++ K+++NP +RE + ++ +Y+ RAE+
Sbjct 9 AIEYVKQAVHEDNAGNYNKAFPLYMNALEYFKTHLKYEKNPKIREAITQKFTEYLRRAEE 68
Query 76 LKQLLRQSSA 85
++ +L + +
Sbjct 69 IRAVLDEGGS 78
> Hs17865802
Length=444
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 46/70 (65%), Gaps = 1/70 (1%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRMDDYIT 71
L+ AI L+++AA +D AG+ EA +LY+ A++ + V K++ Q ++ + + +Y+
Sbjct 8 LQKAIDLASKAAQEDKAGNYEEALQLYQHAVQYFLHVVKYEAQGDKAKQSIRAKCTEYLD 67
Query 72 RAEQLKQLLR 81
RAE+LK+ L+
Sbjct 68 RAEKLKEYLK 77
> SPAC2G11.06
Length=432
Score = 44.3 bits (103), Expect = 6e-05, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITR 72
L AI L A D+A P+A++ Y+ AL+ + + K+++N +E + ++ +Y+ R
Sbjct 7 LSKAISLVKTAIDNDNAEQYPDAYKYYQSALDYFMMALKYEKNEKSKEIIRSKVIEYLDR 66
Query 73 AEQLKQLLRQSS 84
AE+LK L++ +
Sbjct 67 AEKLKVYLQEKN 78
> YPR173c
Length=437
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/78 (28%), Positives = 40/78 (51%), Gaps = 0/78 (0%)
Query 13 LRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITR 72
L I L +A D+A EA+ Y L+ + K+++NP ++ + + +Y+ R
Sbjct 7 LTKGIELVQKAIDLDTATQYEEAYTAYYNGLDYLMLALKYEKNPKSKDLIRAKFTEYLNR 66
Query 73 AEQLKQLLRQSSASHLRQ 90
AEQLK+ L A+ ++
Sbjct 67 AEQLKKHLESEEANAAKK 84
> CE26059
Length=216
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 1/77 (1%)
Query 8 LSDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQ-QNPTLRERLYRRM 66
+S L+ AI L +A +D+AG +A LY +A+E + K++ Q R + ++
Sbjct 1 MSVPALQKAIELVTKATEEDTAGRYDQALRLYDQAIEYFLHAIKYESQGDKQRNAIRDKV 60
Query 67 DDYITRAEQLKQLLRQS 83
Y+ RAEQ+K L+
Sbjct 61 GQYLNRAEQIKTHLKDG 77
> At1g08270
Length=126
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 12/45 (26%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 42 ALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQLKQLLRQSSAS 86
ALE +++ K+++NP +R+ + + +Y+ RAE+++ +L + +
Sbjct 3 ALEYFKIYLKYEKNPRIRDAITDKFYEYLRRAEEIRAVLVEVGSG 47
> 7299160
Length=520
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 9 SDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDD 68
++ L+ AI L +A A D + EA+ LY AL+ + + + + T R L R
Sbjct 272 TEHTLQKAIDLVTQACAYDEKHNYKEAYYLYCSALQYFVPLITEETDATKRLALRNRALS 331
Query 69 YITRAEQLKQLL 80
Y RAE++K +
Sbjct 332 YTKRAEEIKNCI 343
> CE04405
Length=655
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 45/103 (43%), Gaps = 24/103 (23%)
Query 1 LRLMEFGLSDERLRTAIRLSNEAAAK---DSAGHLPE-----------AFELYKRALENW 46
L +M+F LSDE + + ++AAK SA L E FE + + W
Sbjct 549 LYMMQFELSDETGQVYVTAFGDSAAKIVGKSAAELGELHDESPDEYNAIFERLQFVPKMW 608
Query 47 RVVCKFQQ-NPTLRERLY------RRMDDYITRAEQLKQLLRQ 82
R+ CK N +R+++ D YI E LKQ++ Q
Sbjct 609 RLRCKMDSYNEEVRQKMTVYGVDDVNQDKYI---ENLKQMIEQ 648
> Hs20270349
Length=249
Score = 29.3 bits (64), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 23 AAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQLKQLLRQ 82
A DS P+A Y+ ++ V K ++ T R L ++ Y+ RAE +K+ L Q
Sbjct 22 AVELDSESRYPQALVCYQEGIDLLLQVLKGTKDNTKRCNLREKISKYMDRAENIKKYLDQ 81
> CE25622
Length=297
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 21 NEAAAK-DSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQ 75
NEAA +SA + A LY L+ + + + Q+N T +Y R+D T+A Q
Sbjct 155 NEAATSCESADNALLAAPLYPEDLKKFSELGEVQRNKTANRNIYARIDGIRTKACQ 210
> SPBC3F6.01c
Length=473
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 14 RTAIRLSNEAAAKDSAGHLPEAFELYKRALE 44
+ A+ L NEA GH+ +A +LY +A+E
Sbjct 3 KEALELKNEANKFLKEGHIVQAIDLYTKAIE 33
> Hs19923723_2
Length=835
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 35 AFELYKRALENWRVVCKFQQNPTLRERLYRRMDDYITRAEQLKQL 79
A + Y++ ++ + + +PT RE + RR +Y+ RAE + L
Sbjct 31 ASDFYRKGVDLLLEGVQGESSPTRREAVKRRTAEYLMRAESISSL 75
> Hs22048143
Length=1720
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 48/101 (47%), Gaps = 8/101 (7%)
Query 2 RLMEFGLSDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRER 61
R++ G SDE+ LS+ + AG L EAF L AL VV ++NP LR+
Sbjct 142 RVLVEGGSDEKRLCLQLLSDVLRGQGEAGQLEEAFSL---ALLPQLVVSLREENPALRKD 198
Query 62 LYRRMDDYITR--AEQLKQLLRQ---SSASHLRQRLLLYTP 97
+ + + R E L+ L++Q S+ + LR L P
Sbjct 199 ALQILHICLKRSPGEVLRTLIQQGLESTDARLRASTALLLP 239
> Hs7705284
Length=565
Score = 26.9 bits (58), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 29/63 (46%), Gaps = 0/63 (0%)
Query 7 GLSDERLRTAIRLSNEAAAKDSAGHLPEAFELYKRALENWRVVCKFQQNPTLRERLYRRM 66
G + + T L ++ +D+ L +LY RA +N V KF+Q L L + M
Sbjct 215 GDNSRAISTICSLEKKSLLRDNVDLLGSLADLYFRAGDNKNSVLKFEQAQMLDLYLIKGM 274
Query 67 DDY 69
D Y
Sbjct 275 DVY 277
Lambda K H
0.323 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198045380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40