bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2064_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
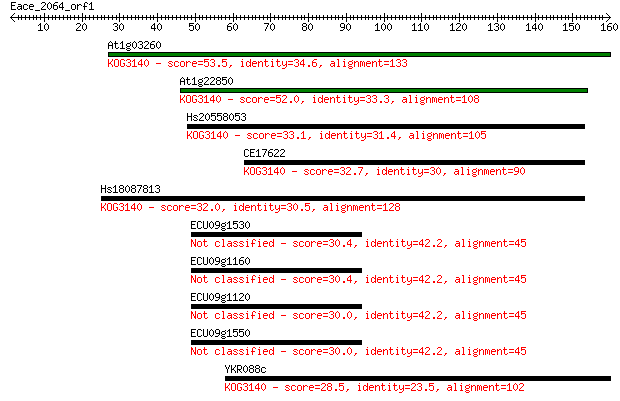
At1g03260 53.5 2e-07
At1g22850 52.0 5e-07
Hs20558053 33.1 0.22
CE17622 32.7 0.29
Hs18087813 32.0 0.57
ECU09g1530 30.4 1.4
ECU09g1160 30.4 1.7
ECU09g1120 30.0 1.9
ECU09g1550 30.0 2.0
YKR088c 28.5 6.1
> At1g03260
Length=269
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 46/138 (33%), Positives = 69/138 (50%), Gaps = 7/138 (5%)
Query 27 FLP-LKDELRQVVE-CQRNSPATYVLTYIVAGLL-IPAPLLSVLAGVLMGPSPLAVVVIL 83
FLP LKD L + E P L YI ++ +PA +L++ G L G P+ V
Sbjct 26 FLPKLKDFLLWIKEDLGPFGPLALALAYIPLTIVAVPASVLTLGGGYLFG-LPVGFVADS 84
Query 84 CGSLGSACLAFAISRFLLRQFVVRRFVRRSQQLQAIELALQKDSVKLVLCTRV--ILPFT 141
G+ A AF + R + + +V + ++ + QA+ +A+QK K+VL RV ILPF
Sbjct 85 LGATLGATAAFLLGRTIGKSYVTSK-IKHYPKFQAVSVAIQKSGFKIVLLLRVVPILPFN 143
Query 142 FNNYFLGTTPISATTFAL 159
NY L TP+ + L
Sbjct 144 MLNYLLSVTPVRLGEYML 161
> At1g22850
Length=344
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 67/123 (54%), Gaps = 17/123 (13%)
Query 46 ATYVLTYIVAG-------------LLIPAPLLSVLAGVLMGPSPLAVVVILCGSLGSACL 92
+TY+ Y AG L IPA L++ AG+L GP ++V + G++ +A +
Sbjct 142 STYIEGYGTAGYALFIAVYAGLEILAIPALPLTMSAGLLFGPLIGTIIVSISGTM-AASV 200
Query 93 AFAISRFLLRQFVVRRFVRRSQQLQAIELALQKDSVKLVLCTRV--ILPFTFNNYFLGTT 150
AF I+R+ R+ ++ + V +++ AI+ A+ ++ ++V R+ +LPF+ NY G T
Sbjct 201 AFLIARYFARERIL-KLVEDNKKFLAIDKAIGENGFRVVTLLRLSPLLPFSLGNYLYGLT 259
Query 151 PIS 153
+
Sbjct 260 SVK 262
> Hs20558053
Length=291
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 60/111 (54%), Gaps = 12/111 (10%)
Query 48 YVLTYI-VAGLLIPAPL-LSVLAGVLMGPSPLAVVVI-LCGSLGSA---CLAFAISRFLL 101
Y TYI + IP + LS+L+G L P PLA+ ++ LC LG++ L++ + R ++
Sbjct 117 YFATYIFLQTFAIPGSIFLSILSGFLY-PFPLALFLVCLCSGLGASFCYMLSYLVGRPVV 175
Query 102 RQFVVRRFVRRSQQLQAIELALQKDSVKLVLCTRVILPFTFNNYFLGTTPI 152
+++ + V+ SQQ++ L + ++ R I PF N + T+P+
Sbjct 176 YKYLTEKAVKWSQQVERHREHL----INYIIFLR-ITPFLPNWFINITSPV 221
> CE17622
Length=246
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 44/92 (47%), Gaps = 18/92 (19%)
Query 63 LLSVLAGVLMGPSPLAVVVILCGSLGSACLAFAISRFLLRQFVVRRFVRRSQQLQAIELA 122
L++L+G L P +A+V++ S A + + IS+ R FV+++F R + Q +L+
Sbjct 91 FLTILSGYLF-PFYVAIVLVCSCSATGAAICYTISKLFGRSFVLQKFPERIAKWQD-DLS 148
Query 123 LQKDSVKLVLCTRVILPFTFNNY--FLGTTPI 152
+D F NY FL TPI
Sbjct 149 KHRDD--------------FLNYMIFLRVTPI 166
> Hs18087813
Length=264
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 39/138 (28%), Positives = 61/138 (44%), Gaps = 12/138 (8%)
Query 25 LWFLPLKDELRQVVEC---QRNSPATYVL-----TYIVA-GLLIP-APLLSVLAGVLMGP 74
LWF ELR++ E R YV Y+ G IP + L+VLAG L GP
Sbjct 42 LWFPSDLAELRELSEVLREYRKEHQAYVFLLFCGAYLYKQGFAIPGSSFLNVLAGALFGP 101
Query 75 SPLAVVVILCGSLGSACLAFAISRFLLRQFVVRRFVRRSQQLQAIELALQKDSVKLVLCT 134
++ + S+G+ C + +S +Q VV F + LQ ++ ++S+ L
Sbjct 102 WLGLLLCCVLTSVGATC-CYLLSSIFGKQLVVSYFPDKVALLQR-KVEENRNSLFFFLLF 159
Query 135 RVILPFTFNNYFLGTTPI 152
+ P T N + + PI
Sbjct 160 LRLFPMTPNWFLNLSAPI 177
> ECU09g1530
Length=570
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 4/47 (8%)
Query 49 VLTYIVAGLLIPAPLLSVLAGVLMGPSPLAVVVILCGS--LGSACLA 93
V + VAG I A +++V GVL G S + V V++CG+ LG+ C+
Sbjct 422 VYSMTVAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 466
> ECU09g1160
Length=534
Score = 30.4 bits (67), Expect = 1.7, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 4/47 (8%)
Query 49 VLTYIVAGLLIPAPLLSVLAGVLMGPSPLAVVVILCGS--LGSACLA 93
V + VAG I A +++V GVL G S + V V++CG+ LG+ C+
Sbjct 412 VYSMTVAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 456
> ECU09g1120
Length=574
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 4/47 (8%)
Query 49 VLTYIVAGLLIPAPLLSVLAGVLMGPSPLAVVVILCGS--LGSACLA 93
V + VAG I A +++V GVL G S + V V++CG+ LG+ C+
Sbjct 452 VYSMTVAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 496
> ECU09g1550
Length=591
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 4/47 (8%)
Query 49 VLTYIVAGLLIPAPLLSVLAGVLMGPSPLAVVVILCGS--LGSACLA 93
V + VAG I A +++V GVL G S + V V++CG+ LG+ C+
Sbjct 466 VYSMTVAG--IGAVMMAVQGGVLCGMSLMQVCVVVCGNLVLGALCVG 510
> YKR088c
Length=337
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 24/105 (22%), Positives = 48/105 (45%), Gaps = 4/105 (3%)
Query 58 LIPAPLLSVLAGVLMGPSPLAVVVILCGSLGSACLAFAISRFLLRQFVVRRFVRRSQQLQ 117
+I LLS G++ G S V + GS+ + +F + + +L + V +++ +
Sbjct 152 MIGYSLLSTTTGLIYGVSFEGWVTLALGSVTGSIASFVVFKTILHS-RAEKLVHLNRRFE 210
Query 118 AIELALQKDSVKLVLCTRVILPFTF---NNYFLGTTPISATTFAL 159
A+ LQ+++ +L + PF + N G IS F++
Sbjct 211 ALASILQENNSYWILALLRLCPFPYSLTNGAIAGVYGISVRNFSI 255
Lambda K H
0.330 0.142 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40