bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2065_orf1
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
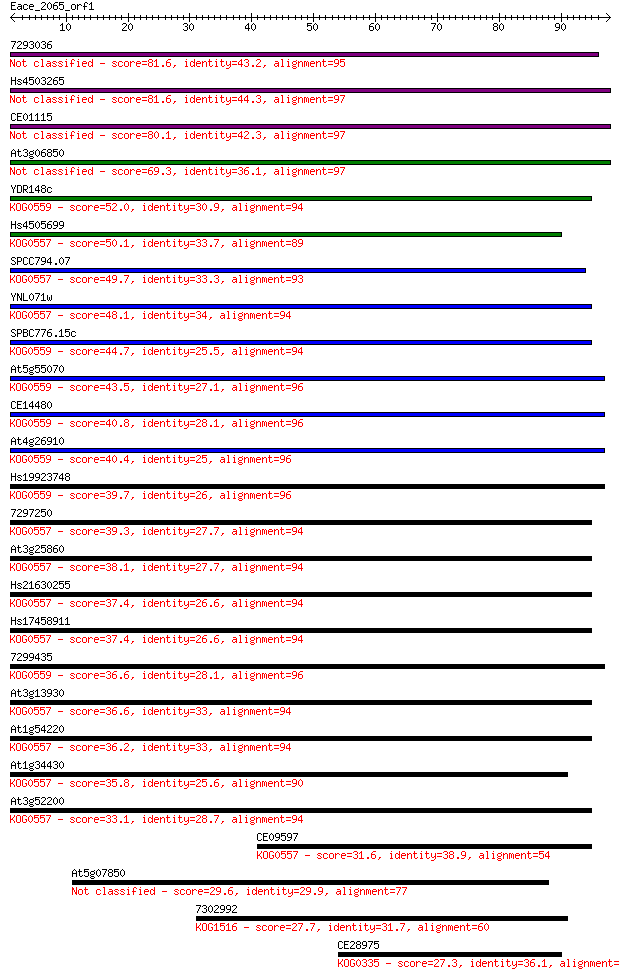
7293036 81.6 3e-16
Hs4503265 81.6 4e-16
CE01115 80.1 1e-15
At3g06850 69.3 2e-12
YDR148c 52.0 3e-07
Hs4505699 50.1 9e-07
SPCC794.07 49.7 1e-06
YNL071w 48.1 4e-06
SPBC776.15c 44.7 4e-05
At5g55070 43.5 9e-05
CE14480 40.8 7e-04
At4g26910 40.4 8e-04
Hs19923748 39.7 0.001
7297250 39.3 0.002
At3g25860 38.1 0.004
Hs21630255 37.4 0.007
Hs17458911 37.4 0.007
7299435 36.6 0.011
At3g13930 36.6 0.013
At1g54220 36.2 0.016
At1g34430 35.8 0.020
At3g52200 33.1 0.12
CE09597 31.6 0.36
At5g07850 29.6 1.6
7302992 27.7 4.9
CE28975 27.3 7.4
> 7293036
Length=462
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 55/95 (57%), Gaps = 8/95 (8%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +LSN+GVI GTY HP + Q I +GR + +PRF D+ +V +++
Sbjct 376 GTFSLSNIGVIGGTYTHPCIMAPQVAIGAMGRTKAVPRFNDKDE--------VVKAYVMS 427
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLH 95
S+SADHR DG T+ FS K LENP + LLH
Sbjct 428 VSWSADHRVIDGVTMASFSNVWKQYLENPALFLLH 462
> Hs4503265
Length=482
Score = 81.6 bits (200), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 43/97 (44%), Positives = 58/97 (59%), Gaps = 8/97 (8%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT LSN+G I GT+ P++ + I +G ++ +PRF + GE A I+N
Sbjct 394 GTFTLSNIGSIGGTFAKPVIMPPEVAIGALGSIKAIPRF----NQKGEVYKA----QIMN 445
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHLK 97
S+SADHR DGAT++RFS K+ LENP MLL LK
Sbjct 446 VSWSADHRVIDGATMSRFSNLWKSYLENPAFMLLDLK 482
> CE01115
Length=448
Score = 80.1 bits (196), Expect = 1e-15, Method: Composition-based stats.
Identities = 41/97 (42%), Positives = 57/97 (58%), Gaps = 8/97 (8%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +LSN+G I GTY P++F Q I +G++ +LPRF D+ ++ +I+
Sbjct 360 GTFSLSNIGNIGGTYASPVVFPPQVAIGAIGKIEKLPRFDKHDN--------VIPVNIMK 411
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHLK 97
S+ ADHR DGAT+ RFS K LE+P ML LK
Sbjct 412 VSWCADHRVVDGATMARFSNRWKFYLEHPSAMLAQLK 448
> At3g06850
Length=483
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 57/97 (58%), Gaps = 8/97 (8%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GTI LSN+G I G + PLL + I+ +GR+ ++P+F + EG+ + I+
Sbjct 395 GTITLSNIGAIGGKFGSPLLNLPEVAIIALGRIEKVPKF------SKEGT--VYPASIMM 446
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHLK 97
+ +ADHR DGAT+ RF K +E PE+++L ++
Sbjct 447 VNIAADHRVLDGATVARFCCQWKEYVEKPELLMLQMR 483
> YDR148c
Length=463
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/94 (30%), Positives = 48/94 (51%), Gaps = 9/94 (9%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +SN GV Y P++ Q ++G+ V++ P V+ +V+R ++
Sbjct 378 GTFTISNGGVFGSLYGTPIINSPQTAVLGLHGVKERPVTVN---------GQIVSRPMMY 428
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ + DHR DG F K++K L+E+P MLL
Sbjct 429 LALTYDHRLLDGREAVTFLKTVKELIEDPRKMLL 462
> Hs4505699
Length=501
Score = 50.1 bits (118), Expect = 9e-07, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 46/89 (51%), Gaps = 4/89 (4%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
G+ ++SN+G+ ++ QA I+ VGR R + + + EG++ L R +I
Sbjct 413 GSFSISNLGMFGIDEFTAVINPPQACILAVGRFRPVLKLTEDE----EGNAKLQQRQLIT 468
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENP 89
+ S+D R D TRF KS K LENP
Sbjct 469 VTMSSDSRVVDDELATRFLKSFKANLENP 497
> SPCC794.07
Length=483
Score = 49.7 bits (117), Expect = 1e-06, Method: Composition-based stats.
Identities = 31/96 (32%), Positives = 48/96 (50%), Gaps = 11/96 (11%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSS--ALVARDI 58
GT +SN+G+ ++ QA I+ VG + D + +S I
Sbjct 396 GTFTISNLGMFPVDQFTAIINPPQACILAVG--------TTVDTVVPDSTSEKGFKVAPI 447
Query 59 INCSFSADHRHCDGATITRFSKSIKTLLENP-EMML 93
+ C+ S+DHR DGA RF+ ++K +LENP E+ML
Sbjct 448 MKCTLSSDHRVVDGAMAARFTTALKKILENPLEIML 483
> YNL071w
Length=482
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 32/95 (33%), Positives = 51/95 (53%), Gaps = 7/95 (7%)
Query 1 GTIALSNVGVISGTYIHPLLFDG-QAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDII 59
GTI +SN+G+ + + + + Q+ I+ + V + V+ +DAA E + + I
Sbjct 394 GTICISNMGMNNAVNMFTSIINPPQSTILAIATVER----VAVEDAAAENGFSFDNQVTI 449
Query 60 NCSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+F DHR DGA F K +KT++ENP MLL
Sbjct 450 TGTF--DHRTIDGAKGAEFMKELKTVIENPLEMLL 482
> SPBC776.15c
Length=452
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 45/94 (47%), Gaps = 9/94 (9%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +SN G+ Y P++ Q ++G+ +++ P ++ +V R ++
Sbjct 367 GTFTISNGGIFGSLYGTPIINLPQTAVLGLHAIKERPVVIN---------GQVVPRPMMY 417
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ + DHR DG F + +K +E+P MLL
Sbjct 418 LALTYDHRMVDGREAVTFLRLVKEYIEDPAKMLL 451
> At5g55070
Length=464
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 47/96 (48%), Gaps = 9/96 (9%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
G+ +SN GV P++ Q+ I+G+ + Q P V ++V R ++
Sbjct 378 GSFTVSNGGVYGSLISTPIINPPQSAILGMHSIVQRPMVVG---------GSVVPRPMMY 428
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHL 96
+ + DHR DG F + IK ++E+P+ +LL +
Sbjct 429 VALTYDHRLIDGREAVYFLRRIKDVVEDPQRLLLDI 464
> CE14480
Length=452
Score = 40.8 bits (94), Expect = 7e-04, Method: Composition-based stats.
Identities = 27/96 (28%), Positives = 42/96 (43%), Gaps = 20/96 (20%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +SN GV + P++ Q+ I+G+ P R I+
Sbjct 377 GTFTISNGGVFGSMFGTPIINPPQSAILGM----HGPEI----------------RPIMQ 416
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHL 96
+ + DHR DG F K IKT +E+P +M ++L
Sbjct 417 IALTYDHRLIDGREAVTFLKKIKTAVEDPRIMFMNL 452
> At4g26910
Length=511
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 9/96 (9%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
G+ +SN GV P++ Q+ I+G+ + P V ++V R ++
Sbjct 425 GSFTVSNGGVYGSLISTPIINPPQSAILGMHSIVSRPMVVG---------GSVVPRPMMY 475
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHL 96
+ + DHR DG F + +K ++E+P+ +LL +
Sbjct 476 VALTYDHRLIDGREAVYFLRRVKDVVEDPQRLLLDI 511
> Hs19923748
Length=453
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 44/96 (45%), Gaps = 9/96 (9%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +SN GV + P++ Q+ I+G+ + P + + R ++
Sbjct 367 GTFTISNGGVFGSLFGTPIINPPQSAILGMHGIFDRPVAIG---------GKVEVRPMMY 417
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHL 96
+ + DHR DG F + IK +E+P ++LL L
Sbjct 418 VALTYDHRLIDGREAVTFLRKIKAAVEDPRVLLLDL 453
> 7297250
Length=512
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 49/95 (51%), Gaps = 8/95 (8%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRV-RQLPRFVSCDDAAGEGSSALVARDII 59
GTI++SN+G+ ++ Q+ I+ +G +QL D + +G + +++
Sbjct 425 GTISVSNLGMFGVNQFAAVINPPQSCILAIGTTTKQL----VADPDSLKGFKEV---NML 477
Query 60 NCSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ SADHR DGA R+ + + +E+P M+L
Sbjct 478 TVTLSADHRVVDGAVAARWLQHFRDYMEDPSNMVL 512
> At3g25860
Length=480
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 26/94 (27%), Positives = 44/94 (46%), Gaps = 8/94 (8%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT LSN+G+ +L GQ I+ VG + P V+ D ++ +
Sbjct 395 GTFTLSNLGMFGVDRFDAILPPGQGAIMAVGASK--PTVVADKDGF------FSVKNTML 446
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ +ADHR GA + F ++ ++ENP+ + L
Sbjct 447 VNVTADHRIVYGADLAAFLQTFAKIIENPDSLTL 480
> Hs21630255
Length=613
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 43/94 (45%), Gaps = 7/94 (7%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +SN+G+ ++ QA I+ +G + V D+ G +++++
Sbjct 527 GTFTISNLGMFGIKNFSAIINPPQACILAIGASED--KLVPADNEKGFDVASMMS----- 579
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ S DHR DGA ++ + LE P MLL
Sbjct 580 VTLSCDHRVVDGAVGAQWLAEFRKYLEKPITMLL 613
> Hs17458911
Length=647
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 43/94 (45%), Gaps = 7/94 (7%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +SN+G+ ++ QA I+ +G + V D+ G +++++
Sbjct 561 GTFTISNLGMFGIKNFSAIINPPQACILAIGASED--KLVPADNEKGFDVASMMS----- 613
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ S DHR DGA ++ + LE P MLL
Sbjct 614 VTLSCDHRVVDGAVGAQWLAEFRKYLEKPITMLL 647
> 7299435
Length=468
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 45/96 (46%), Gaps = 9/96 (9%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT +SN GV P++ Q+ I+G+ + + P V GE + R ++
Sbjct 382 GTFTISNGGVFGSLMGTPIINPPQSAILGMHGIFERPIAVK-----GE----VKIRPMMY 432
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLLHL 96
+ + DHR DG F + IK +ENP +++ L
Sbjct 433 IALTYDHRIIDGREAVLFLRKIKAAVENPAIIVAGL 468
> At3g13930
Length=508
Score = 36.6 bits (83), Expect = 0.013, Method: Composition-based stats.
Identities = 31/99 (31%), Positives = 43/99 (43%), Gaps = 16/99 (16%)
Query 1 GTIALSNVGVISG-----TYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVA 55
GT +SN+G G I+P QA I+ +G + R V G G
Sbjct 421 GTFTVSNLGGPFGIKQFCAVINP----PQAAILAIGSAEK--RVVP-----GTGPDQYNV 469
Query 56 RDIINCSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
++ + S DHR DGA + K+ K +E PE MLL
Sbjct 470 ASYMSVTLSCDHRVIDGAIGAEWLKAFKGYIETPESMLL 508
> At1g54220
Length=424
Score = 36.2 bits (82), Expect = 0.016, Method: Composition-based stats.
Identities = 31/99 (31%), Positives = 43/99 (43%), Gaps = 16/99 (16%)
Query 1 GTIALSNVGVISG-----TYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVA 55
GT +SN+G G ++P QA I+ VG + R V G G
Sbjct 337 GTFTVSNLGGPFGIKQFCAVVNP----PQAAILAVGSAEK--RVVP-----GNGPDQFNF 385
Query 56 RDIINCSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ + S DHR DGA + K+ K +ENP+ MLL
Sbjct 386 ASYMPVTLSCDHRVVDGAIGAEWLKAFKGYIENPKSMLL 424
> At1g34430
Length=465
Score = 35.8 bits (81), Expect = 0.020, Method: Composition-based stats.
Identities = 23/90 (25%), Positives = 45/90 (50%), Gaps = 8/90 (8%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT LSN+G+ +L G I+ VG + P V+ D + ++ +
Sbjct 380 GTFTLSNLGMFGVDRFDAILPPGTGAIMAVGASQ--PSVVATKDGR------IGMKNQMQ 431
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPE 90
+ +ADHR GA + +F +++ +++E+P+
Sbjct 432 VNVTADHRVIYGADLAQFLQTLASIIEDPK 461
> At3g52200
Length=637
Score = 33.1 bits (74), Expect = 0.12, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query 1 GTIALSNVGVISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEGSSALVARDIIN 60
GT ++SN+G+ ++ QA I+ VGR ++ V D G ++V + +N
Sbjct 548 GTFSISNLGMYPVDNFCAIINPPQAGILAVGRGNKVVEPVIGLD--GIEKPSVVTK--MN 603
Query 61 CSFSADHRHCDGATITRFSKSIKTLLENPEMMLL 94
+ SADHR DG F +++ E+ +LL
Sbjct 604 VTLSADHRIFDGQVGASFMSELRSNFEDVRRLLL 637
> CE09597
Length=507
Score = 31.6 bits (70), Expect = 0.36, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 26/62 (41%), Gaps = 8/62 (12%)
Query 41 SCDDAAGEGSSALVARD--------IINCSFSADHRHCDGATITRFSKSIKTLLENPEMM 92
SC A G S LV + + + S DHR DGA + + K LE P M
Sbjct 446 SCILAIGGASDKLVPDEAEGYKKIKTMKVTLSCDHRTVDGAVGAVWLRHFKEFLEKPHTM 505
Query 93 LL 94
LL
Sbjct 506 LL 507
> At5g07850
Length=456
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 23/91 (25%), Positives = 38/91 (41%), Gaps = 14/91 (15%)
Query 11 ISGTYIHPLLFDGQAVIVGVGRVRQLPRFVSCDDAAGEG----SSALVARDIINCSFSA- 65
++ T +H G+ + R FV C+D+ G G S L RDI+ ++
Sbjct 74 LAETLVHFYPLAGRLSTLKTDNPRSYSVFVDCNDSPGAGFIHAKSDLSVRDIVGSNYVPL 133
Query 66 ------DHRHC---DGATITRFSKSIKTLLE 87
DH DG T++ FS + L++
Sbjct 134 VVQSFFDHHKAVSHDGHTMSLFSVKVTELVD 164
> 7302992
Length=507
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 26/60 (43%), Gaps = 8/60 (13%)
Query 31 GRVRQLPRFVSCDDAAGEGSSALVARDIINCSFSADHRHCDGATITRFSKSIKTLLENPE 90
GR+ Q+P +S AGEG S + R IN A+ F + + LL PE
Sbjct 325 GRINQMPWILSLSSRAGEG-SLFIMRAFINPKLRAEFNE-------NFLEHMALLLNLPE 376
> CE28975
Length=520
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 54 VARDIINCSFSADHRHCDGATITRFSKSIKTLLENP 89
V R++ +C FSA H + +I S+ + L ENP
Sbjct 279 VLRNLQSCFFSASHNRNEDGSIALTSRQSRMLGENP 314
Lambda K H
0.324 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198045380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40