bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
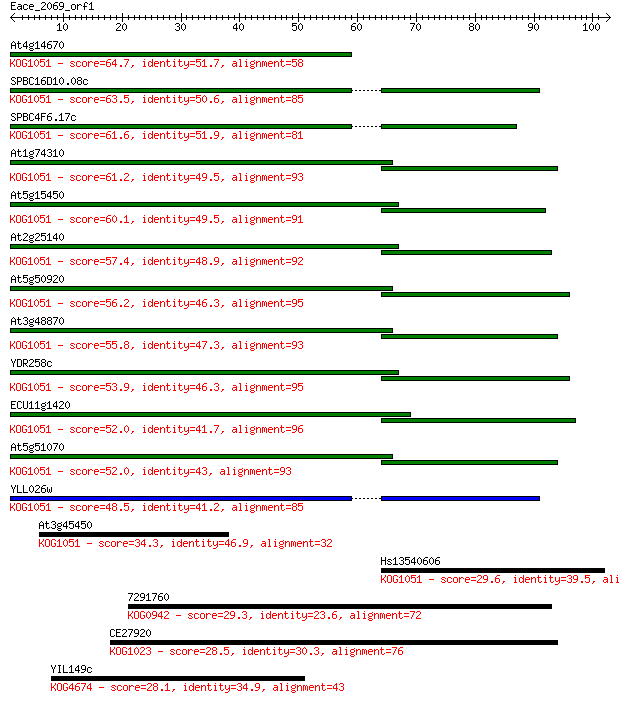
Query= Eace_2069_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
At4g14670 64.7 4e-11
SPBC16D10.08c 63.5 8e-11
SPBC4F6.17c 61.6 4e-10
At1g74310 61.2 4e-10
At5g15450 60.1 1e-09
At2g25140 57.4 6e-09
At5g50920 56.2 1e-08
At3g48870 55.8 2e-08
YDR258c 53.9 7e-08
ECU11g1420 52.0 3e-07
At5g51070 52.0 3e-07
YLL026w 48.5 3e-06
At3g45450 34.3 0.054
Hs13540606 29.6 1.3
7291760 29.3 2.1
CE27920 28.5 3.2
YIL149c 28.1 3.9
> At4g14670
Length=623
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 30/58 (51%), Positives = 38/58 (65%), Gaps = 0/58 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL 58
ATTLEEY+ H+EKDAAF RRFQ + V PS +SIL+ +K YE + V + D L
Sbjct 279 ATTLEEYRTHVEKDAAFERRFQQVFVAEPSVPDTISILRGLKEKYEGHHGVRIQDRAL 336
> SPBC16D10.08c
Length=905
Score = 63.5 bits (153), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 33/58 (56%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL 58
ATTL EYK +IEKDAAF RRFQ I+V+ PS E +SIL+ +K YE + V +SD L
Sbjct 317 ATTLAEYKKYIEKDAAFERRFQIILVKEPSIEDTISILRGLKEKYEVHHGVTISDRAL 374
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHI 90
++ +G VD N +II+TSN+G +++
Sbjct 715 ITSGQGQVVDAKNAVIIMTSNLGAEYL 741
> SPBC4F6.17c
Length=803
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 40/58 (68%), Gaps = 0/58 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL 58
ATTLEEY+ +IEKDAA RRFQ ++V PS +SIL+ +K YE + V ++D+ L
Sbjct 246 ATTLEEYRKYIEKDAALARRFQAVMVNEPSVADTISILRGLKERYEVHHGVRITDDAL 303
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIG 86
L+DS+G +VDF + LI++TSN+G
Sbjct 633 LTDSQGRKVDFRSTLIVMTSNLG 655
> At1g74310
Length=911
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTLEEY+ ++EKDAAF RRFQ + V PS +SIL+ +K YE + V + D L
Sbjct 314 ATTLEEYRKYVEKDAAFERRFQQVYVAEPSVPDTISILRGLKEKYEGHHGVRIQDRALIN 373
Query 61 IVALS 65
LS
Sbjct 374 AAQLS 378
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 23/30 (76%), Gaps = 0/30 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEG 93
L+D +G VDF N +II+TSN+G +H+L G
Sbjct 700 LTDGQGRTVDFRNSVIIMTSNLGAEHLLAG 729
> At5g15450
Length=968
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ +IEKD A RRFQ + V+ P+ E +SIL+ ++ YE + V +SD L
Sbjct 389 ATTLDEYRKYIEKDPALERRFQQVYVDQPTVEDTISILRGLRERYELHHGVRISDSALVE 448
Query 61 IVALSD 66
LSD
Sbjct 449 AAILSD 454
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHIL 91
++DS+G V FTN +II+TSN+G Q IL
Sbjct 779 VTDSQGRTVSFTNTVIIMTSNVGSQFIL 806
> At2g25140
Length=874
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL EY+ +IEKD A RRFQ ++ PS E +SIL+ ++ YE + V +SD L
Sbjct 304 ATTLTEYRKYIEKDPALERRFQQVLCVQPSVEDTISILRGLRERYELHHGVTISDSALVS 363
Query 61 IVALSD 66
L+D
Sbjct 364 AAVLAD 369
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 17/29 (58%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILE 92
++DS+G V F NC++I+TSNIG HILE
Sbjct 694 ITDSQGRTVSFKNCVVIMTSNIGSHHILE 722
> At5g50920
Length=929
Score = 56.2 bits (134), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+EY+ HIEKD A RRFQ + V P+ + + ILK ++ YE + + +DE L
Sbjct 408 ATTLDEYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDESLVA 467
Query 61 IVALS 65
LS
Sbjct 468 AAQLS 472
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYK 95
L+DSKG VDF N L+I+TSN+G I +G +
Sbjct 739 LTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGR 770
> At3g48870
Length=952
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATT++EY+ HIEKD A RRFQ + V P+ E A+ IL+ ++ YE + + +DE L
Sbjct 429 ATTIDEYRKHIEKDPALERRFQPVKVPEPTVEEAIQILQGLRERYEIHHKLRYTDEALVA 488
Query 61 IVALS 65
LS
Sbjct 489 AAQLS 493
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 17/30 (56%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEG 93
L+DSKG VDF N L+I+TSN+G I +G
Sbjct 760 LTDSKGRTVDFKNTLLIMTSNVGSSVIEKG 789
> YDR258c
Length=811
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 41/66 (62%), Gaps = 1/66 (1%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATTL+E+K+ IEKD A RRFQ I++ PS +SIL+ +K YE + V ++D L
Sbjct 249 ATTLDEFKI-IEKDPALSRRFQPILLNEPSVSDTISILRGLKERYEVHHGVRITDTALVS 307
Query 61 IVALSD 66
LS+
Sbjct 308 AAVLSN 313
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYK 95
L+DS G+ VDF N +I++TSNIGQ +L K
Sbjct 635 LTDSLGHHVDFRNTIIVMTSNIGQDILLNDTK 666
> ECU11g1420
Length=851
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 44/68 (64%), Gaps = 2/68 (2%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
ATT +EY+ +IE D AF RRF +VV PS E ++++L+ +K E ++ V ++D +
Sbjct 300 ATTHDEYRKYIENDPAFERRFVQVVVNEPSIEDSITMLRGLKGRLEAYHGVKIADSAI-- 357
Query 61 IVALSDSK 68
+ A++ SK
Sbjct 358 VYAVNSSK 365
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEGYKE 96
++D KG VDF NC++I+TSN+GQ+ I+ E
Sbjct 683 VTDGKGAVVDFRNCVVIMTSNLGQEIIMRSRGE 715
> At5g51070
Length=945
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEG 60
+TTL+E++ EKD A RRFQ +++ PS+E A+ IL ++ YE ++ + E ++
Sbjct 427 STTLDEFRSQFEKDKALARRFQPVLINEPSEEDAVKILLGLREKYEAHHNCKYTMEAIDA 486
Query 61 IVALS 65
V LS
Sbjct 487 AVYLS 491
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 17/30 (56%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHILEG 93
L+DS+G RV F N LII+TSN+G I +G
Sbjct 758 LTDSQGRRVSFKNALIIMTSNVGSLAIAKG 787
> YLL026w
Length=908
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 1 ATTLEEYKLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVL 58
ATT EY+ +EKD AF RRFQ I V PS + ++IL+ ++ YE + V + D L
Sbjct 315 ATTNNEYRSIVEKDGAFERRFQKIEVAEPSVRQTVAILRGLQPKYEIHHGVRILDSAL 372
Score = 31.6 bits (70), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQQHI 90
++ +G +D +NC++I+TSN+G + I
Sbjct 708 ITSGQGKTIDCSNCIVIMTSNLGAEFI 734
> At3g45450
Length=341
Score = 34.3 bits (77), Expect = 0.054, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 6 EYKLHIEKDAAFCRRFQNIVVEAPSKERALSI 37
+Y+ HIE D A RRFQ + V P+ E A+ I
Sbjct 278 QYRKHIENDPALERRFQPVKVPEPTVEEAIQI 309
> Hs13540606
Length=707
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 24/42 (57%), Gaps = 4/42 (9%)
Query 64 LSDSKGNRVDFTNCLIILTSNIGQ----QHILEGYKEARALS 101
L+D KG +D + + I+TSN+ QH L+ +EA +S
Sbjct 476 LTDGKGKTIDCKDAIFIMTSNVASDEIAQHALQLRQEALEMS 517
> 7291760
Length=1122
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 17/72 (23%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 21 FQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEGIVALSDSKGNRVDFTNCLII 80
++N++VE P E L+ L ++ + L E+ ++ L D G+ + +
Sbjct 870 YENLLVELPLAEFFLTKLAGKYSDVDIHQLASLDPELYRNLLYLKDYSGDVSELNLDFTV 929
Query 81 LTSNIGQQHILE 92
+S++GQ I+E
Sbjct 930 ASSSLGQTQIVE 941
> CE27920
Length=1280
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 23/85 (27%), Positives = 40/85 (47%), Gaps = 10/85 (11%)
Query 18 CRRFQNIVVEAPSKERALSILKKVKTNYEKFYSVDLSDEVLEGIVALSDSK----GNRVD 73
C+ +V E SK IL+ + Y D++++G+V + DS+ GN +
Sbjct 735 CQYSVTVVREYCSKGSLHDILRNENLKLDHMYVASFVDDLVKGMVYIHDSELKMHGN-LK 793
Query 74 FTNCLI-----ILTSNIGQQHILEG 93
TNCLI + ++ G + + EG
Sbjct 794 STNCLITSRWTLQIADFGLRELREG 818
> YIL149c
Length=1679
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 8 KLHIEKDAAFCRRFQNIVVEAPSKERALSILKKVKTNYEKFYS 50
KL +EK C+R QNIV++ +E A V K +S
Sbjct 323 KLRLEKSKNECQRLQNIVMDCTKEEEATMTTSAVSPTVGKLFS 365
Lambda K H
0.317 0.132 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181107380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40