bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2100_orf1
Length=96
Score E
Sequences producing significant alignments: (Bits) Value
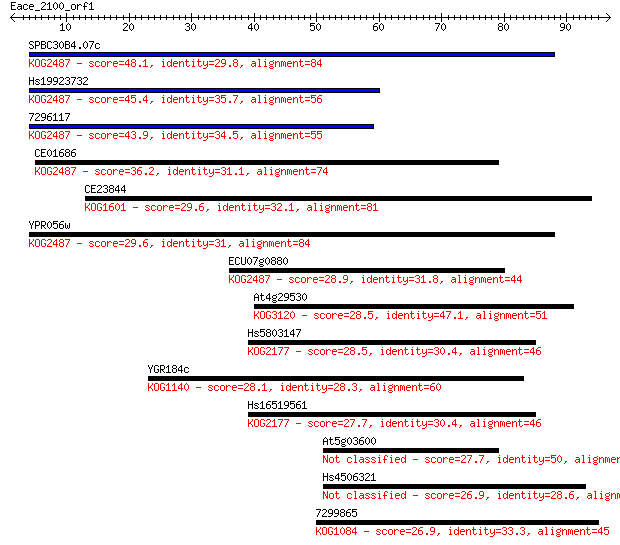
SPBC30B4.07c 48.1 4e-06
Hs19923732 45.4 3e-05
7296117 43.9 8e-05
CE01686 36.2 0.016
CE23844 29.6 1.4
YPR056w 29.6 1.6
ECU07g0880 28.9 2.7
At4g29530 28.5 3.0
Hs5803147 28.5 3.5
YGR184c 28.1 4.6
Hs16519561 27.7 5.6
At5g03600 27.7 5.7
Hs4506321 26.9 8.2
7299865 26.9 8.5
> SPBC30B4.07c
Length=297
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 40/84 (47%), Gaps = 7/84 (8%)
Query 4 ALLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETSD 63
LLQ L+ P +LR + + + CFCH +D+ F+CS CL+I+C
Sbjct 219 GLLQYLMMSLFPDQNLRKHLNTPNQANVDFRATCFCHKKVLDIGFVCSVCLSIFC----- 273
Query 64 NGKERISCDVCKSRFAKGLLKAKL 87
+ R+ C C ++F + KL
Sbjct 274 --EPRVHCSTCHTKFTVEPMSKKL 295
> Hs19923732
Length=308
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 4 ALLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCS 59
+LLQ L++ FLP R + H CFCH N +++ ++CS CL+I+C+
Sbjct 222 SLLQYLLWVFLPDQDQRSQLILPPPVHVDYRAACFCHRNLIEIGYVCSVCLSIFCN 277
> 7296117
Length=276
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 0/55 (0%)
Query 4 ALLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYC 58
LLQ L++ FLP P +R + CFCH +D+ ++CS CL+++C
Sbjct 212 GLLQYLLWVFLPAPQIRHKLVLPPPPKVDYRASCFCHRELIDIGYVCSVCLSVFC 266
> CE01686
Length=345
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 7/74 (9%)
Query 5 LLQLLVFWFLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETSDN 64
LL++L+ L P+ R + +S + C CHH V ++CS CL++ C T
Sbjct 262 LLKILMTNMLTDPTHRAVFSKLSHNSVDYRASCACHHQLVSSGWVCSICLSVLCQYTP-- 319
Query 65 GKERISCDVCKSRF 78
C VCK+ F
Sbjct 320 -----ICKVCKAAF 328
> CE23844
Length=226
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 31/81 (38%), Gaps = 7/81 (8%)
Query 13 FLPPPSLRPSVAAVSIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSETSDNGKERISCD 72
F PPP P VA K A V CH N ICS C + NG+ + C+
Sbjct 123 FTPPPQ-DPLVAEPKPMKKRMAAVQ-CHQN-----SICSNCKTRETTLWRRNGEGGVECN 175
Query 73 VCKSRFAKGLLKAKLAASVDA 93
C F K K L+ D
Sbjct 176 ACNLYFRKNNRKRPLSLRKDG 196
> YPR056w
Length=338
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 38/87 (43%), Gaps = 12/87 (13%)
Query 4 ALLQLLVFWFLPPPSLRPSVAAV---SIHHKTNAGVCFCHHNPVDLCFICSCCLAIYCSE 60
L+Q L PSLRP + S+ +T+ C+ V + FICS CL +
Sbjct 243 GLIQYLATAMFIDPSLRPIIVKPNHGSVDFRTS---CYLTGRVVAVGFICSVCLCVLSII 299
Query 61 TSDNGKERISCDVCKSRFAKGLLKAKL 87
N C C S+F + ++ AKL
Sbjct 300 PPGN-----KCPACDSQFDEHVI-AKL 320
> ECU07g0880
Length=213
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 22/44 (50%), Gaps = 7/44 (15%)
Query 36 VCFCHHNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFA 79
C+CH+ V L +C CL+++C C CKS+F+
Sbjct 171 TCYCHNRQVLLGLVCPICLSVFCRFVP-------VCKKCKSKFS 207
> At4g29530
Length=245
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 29/64 (45%), Gaps = 19/64 (29%)
Query 40 HHNPVDLCFICSCCLAIYCSETS--DNGKERI-----------SCDVCKSRFAKGLLKAK 86
HH+ VD C IY + TS DNG RI SC++C S KGL+
Sbjct 107 HHDLVD------CFSEIYTNPTSLDDNGNLRILPYHSDALPPHSCNLCPSNLCKGLVMDH 160
Query 87 LAAS 90
L AS
Sbjct 161 LRAS 164
> Hs5803147
Length=481
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 19/46 (41%), Gaps = 6/46 (13%)
Query 39 CHHNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLK 84
C HN C CL YC + +E +C +CK F G +
Sbjct 31 CGHN------FCRACLTRYCEIPGPDLEESPTCPLCKEPFRPGSFR 70
> YGR184c
Length=1950
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 26/64 (40%), Gaps = 5/64 (7%)
Query 23 VAAVSIHHKTNAGVCFCHHNPVDLCFIC----SCCLAIYCSETSDNGKERISCDVCKSRF 78
+ V H N G F P+ C C +C L I+C D+ + D+C + F
Sbjct 112 IGDVHKHTGRNCGRKFKIGEPLYRCHECGCDDTCVLCIHCFNPKDHVNHHVCTDIC-TEF 170
Query 79 AKGL 82
G+
Sbjct 171 TSGI 174
> Hs16519561
Length=395
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 19/46 (41%), Gaps = 6/46 (13%)
Query 39 CHHNPVDLCFICSCCLAIYCSETSDNGKERISCDVCKSRFAKGLLK 84
C HN C CL YC + +E +C +CK F G +
Sbjct 31 CGHN------FCRACLTRYCEIPGPDLEESPTCPLCKEPFRPGSFR 70
> At5g03600
Length=322
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 15/28 (53%), Gaps = 1/28 (3%)
Query 51 SCCLAIYCSETSDNGKERISCDVCKSRF 78
SCC YC +SD GK CD KS F
Sbjct 266 SCCEG-YCGRSSDGGKLYTLCDDPKSFF 292
> Hs4506321
Length=979
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 12/42 (28%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 51 SCCLAIYCSETSDNGKERISCDVCKSRFAKGLLKAKLAASVD 92
SC + ++CS+ + I D+ +R AKG+ + +AA+++
Sbjct 902 SCPIIVHCSDGAGRTGTYILIDMVLNRMAKGVKEIDIAATLE 943
> 7299865
Length=3085
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query 50 CSCCLAIYCSETSDNGKER--ISCDVCKSRFAKGLLKAKLAASVDAS 94
CS C A+ S+ ++ ++CDVC+ F+K K+ A S A+
Sbjct 735 CSICSAVVSSKEVTQARKYGVVACDVCRKFFSKMTKKSISANSSTAN 781
Lambda K H
0.327 0.138 0.462
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1201432980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40