bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
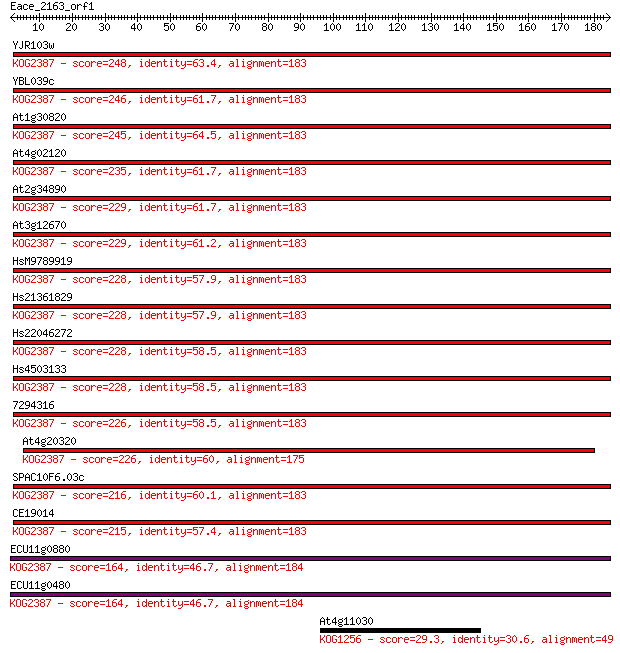
Query= Eace_2163_orf1
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
YJR103w 248 3e-66
YBL039c 246 1e-65
At1g30820 245 3e-65
At4g02120 235 3e-62
At2g34890 229 2e-60
At3g12670 229 2e-60
HsM9789919 228 5e-60
Hs21361829 228 5e-60
Hs22046272 228 5e-60
Hs4503133 228 5e-60
7294316 226 2e-59
At4g20320 226 3e-59
SPAC10F6.03c 216 2e-56
CE19014 215 4e-56
ECU11g0880 164 8e-41
ECU11g0480 164 8e-41
At4g11030 29.3 4.9
> YJR103w
Length=564
Score = 248 bits (634), Expect = 3e-66, Method: Compositional matrix adjust.
Identities = 116/184 (63%), Positives = 151/184 (82%), Gaps = 2/184 (1%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SS G++L++LG++V++IKIDPY+N DAGTMSP EHGECFVLDDGGE DLD
Sbjct 11 ISGIGKGVLASSTGMLLKTLGLKVTSIKIDPYMNIDAGTMSPLEHGECFVLDDGGETDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYER+LG++LSR HNIT+GK+YS VIS+ER+G +LGK VQI+PH+T AIQ W+ RV+
Sbjct 71 LGNYERYLGITLSRDHNITTGKIYSHVISRERRGDYLGKTVQIVPHLTNAIQDWIQRVSK 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL-GE 180
PV T + P+VC+IE+GGTVGDIESA ++EA++Q +GR+ L HV+ VP + GE
Sbjct 131 IPVDDTGLE-PDVCIIELGGTVGDIESAPFVEALRQFQFEVGRENFALIHVSLVPVIHGE 189
Query 181 QKTK 184
QKTK
Sbjct 190 QKTK 193
> YBL039c
Length=579
Score = 246 bits (629), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 113/184 (61%), Positives = 151/184 (82%), Gaps = 2/184 (1%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SS G+++++LG++V++IKIDPY+N DAGTMSP EHGECFVLDDGGE DLD
Sbjct 11 ISGIGKGVLASSTGMLMKTLGLKVTSIKIDPYMNIDAGTMSPLEHGECFVLDDGGETDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYER+LGV+L++ HNIT+GK+YS VI++ER+G +LGK VQI+PH+T AIQ W+ RVA
Sbjct 71 LGNYERYLGVTLTKDHNITTGKIYSHVIAKERKGDYLGKTVQIVPHLTNAIQDWIERVAK 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL-GE 180
PV T + P+VC+IE+GGTVGDIESA ++EA++Q +G++ L HV+ VP + GE
Sbjct 131 IPVDDTGME-PDVCIIELGGTVGDIESAPFVEALRQFQFKVGKENFALIHVSLVPVIHGE 189
Query 181 QKTK 184
QKTK
Sbjct 190 QKTK 193
> At1g30820
Length=600
Score = 245 bits (626), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 118/186 (63%), Positives = 148/186 (79%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SGLGKG T SSIGV+L++ G+RV++IKIDPYLN+DAGTMSP+EHGE FVLDDGGEVDLD
Sbjct 11 VSGLGKGVTASSIGVLLKACGLRVTSIKIDPYLNTDAGTMSPFEHGEVFVLDDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL + L+R +NIT+GK+Y VI++ER+G +LGK VQ++PHVT AIQ W+ RVA
Sbjct 71 LGNYERFLDIKLTRDNNITTGKIYQHVIAKERKGDYLGKTVQVVPHVTDAIQDWIERVAV 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL--- 178
PV D +VC+IE+GGT+GDIESA ++EA+ Q S +G CL HV+ VP L
Sbjct 131 IPV-DGEEDPADVCVIELGGTIGDIESAPFIEALGQFSYRVGPGNFCLVHVSLVPVLNVV 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> At4g02120
Length=539
Score = 235 bits (600), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 113/186 (60%), Positives = 148/186 (79%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SGLGKG T SSIGVVL++ G+ V++IKIDPYLN+DAGTMSP+EHGE FVLDDGGEVDLD
Sbjct 11 VSGLGKGVTASSIGVVLKACGLGVTSIKIDPYLNTDAGTMSPFEHGEVFVLDDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL V+L++ +NIT+GK+Y V+ +ER+G +LGK VQ++PH+T AI+ W+ V+
Sbjct 71 LGNYERFLDVTLTKDNNITTGKIYQSVLDKERKGDYLGKTVQVVPHITDAIKDWIESVSL 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL--- 178
PV +VC+IE+GGTVGDIES ++EA++QLS ++G D CL HV+ +P L
Sbjct 131 IPVDGKEGQA-DVCVIELGGTVGDIESMPFIEALRQLSFSVGPDNFCLIHVSLIPVLGVV 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> At2g34890
Length=597
Score = 229 bits (585), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 113/186 (60%), Positives = 142/186 (76%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SGLGKG T SS+GV+L+S G+RV+ IKIDPYLN DAGT+SPYEHGE FVLDDG EVDLD
Sbjct 11 VSGLGKGITASSVGVLLQSCGLRVTCIKIDPYLNYDAGTISPYEHGEVFVLDDGSEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL +L+R +NIT GK+ V+ +ER+G +LG+ VQI+PHVT I++WV RVA
Sbjct 71 LGNYERFLDTTLTRDNNITYGKIQRYVMEKERKGDYLGETVQIVPHVTDTIREWVERVAM 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL--- 178
PV P+VC+IE+GGT+GD ES + +A+ QLS ++GR+ CL HV VP L
Sbjct 131 IPV-DGKEGPPDVCIIELGGTIGDNESRPFADALSQLSYSVGRENFCLIHVTLVPVLSVV 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> At3g12670
Length=591
Score = 229 bits (585), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 112/186 (60%), Positives = 145/186 (77%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SGLGKG T SSIG++L++ G+RV++IKIDPYLN+DAGTMSP+EHGE FVLDDGGEVDLD
Sbjct 11 VSGLGKGVTASSIGLLLQACGLRVTSIKIDPYLNTDAGTMSPFEHGEVFVLDDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL +L+R +NIT+GK+Y VI +ER+G +LG+ VQ++PHVT AIQ+W+ RVA
Sbjct 71 LGNYERFLDSTLTRDNNITTGKIYQSVIDKERKGDYLGRTVQVVPHVTDAIQEWIERVAN 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAH---VAFVPCL 178
PV P+VC+IE+GGT+GDIES ++EA+ Q S +G CL H V + +
Sbjct 131 VPV-DGKEGPPDVCVIELGGTIGDIESMPFIEALGQFSYKVGPGNFCLVHVSLVPVLSVV 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> HsM9789919
Length=586
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 106/186 (56%), Positives = 141/186 (75%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SSIG +L+S G+RV+AIKIDPY+N DAGT SPYEHGE FVL+DGGEVDLD
Sbjct 11 ISGIGKGIIASSIGTILKSCGLRVTAIKIDPYINIDAGTFSPYEHGEVFVLNDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL ++L + +NIT+GK+Y VI++ER+G +LGK VQ++PH+T A+Q+WV+ A
Sbjct 71 LGNYERFLDINLYKDNNITTGKIYQHVINKERRGDYLGKTVQVVPHITDAVQEWVMNQAK 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL--- 178
PV + P++C+IE+GGT+GDIE ++EA +Q R+ C HV+ VP L
Sbjct 131 VPV-DGNKEEPQICVIELGGTIGDIEGMPFVEAFRQFQFKAKRENFCNIHVSLVPQLSAT 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> Hs21361829
Length=586
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 106/186 (56%), Positives = 141/186 (75%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SSIG +L+S G+RV+AIKIDPY+N DAGT SPYEHGE FVL+DGGEVDLD
Sbjct 11 ISGIGKGIIASSIGTILKSCGLRVTAIKIDPYINIDAGTFSPYEHGEVFVLNDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL ++L + +NIT+GK+Y VI++ER+G +LGK VQ++PH+T A+Q+WV+ A
Sbjct 71 LGNYERFLDINLYKDNNITTGKIYQHVINKERRGDYLGKTVQVVPHITDAVQEWVMNQAK 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL--- 178
PV + P++C+IE+GGT+GDIE ++EA +Q R+ C HV+ VP L
Sbjct 131 VPV-DGNKEEPQICVIELGGTIGDIEGMPFVEAFRQFQFKAKRENFCNIHVSLVPQLSAT 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> Hs22046272
Length=591
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 107/186 (57%), Positives = 142/186 (76%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SS+G +L+S G+ V++IKIDPY+N DAGT SPYEHGE FVLDDGGEVDLD
Sbjct 11 ISGIGKGIIASSVGTILKSCGLHVTSIKIDPYINIDAGTFSPYEHGEVFVLDDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL + L++ +N+T+GK+Y VI++ER+G +LGK VQ++PH+T AIQ+WV+R A
Sbjct 71 LGNYERFLDIRLTKDNNLTTGKIYQYVINKERKGDYLGKTVQVVPHITDAIQEWVMRQAL 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVP---CL 178
PV + P+VC+IE+GGTVGDIES ++EA +Q + R+ C HV+ VP
Sbjct 131 IPVDEDGLE-PQVCVIELGGTVGDIESMPFIEAFRQFQFKVKRENFCNIHVSLVPQPSST 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> Hs4503133
Length=591
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 107/186 (57%), Positives = 142/186 (76%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SS+G +L+S G+ V++IKIDPY+N DAGT SPYEHGE FVLDDGGEVDLD
Sbjct 11 ISGIGKGIIASSVGTILKSCGLHVTSIKIDPYINIDAGTFSPYEHGEVFVLDDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL + L++ +N+T+GK+Y VI++ER+G +LGK VQ++PH+T AIQ+WV+R A
Sbjct 71 LGNYERFLDIRLTKDNNLTTGKIYQYVINKERKGDYLGKTVQVVPHITDAIQEWVMRQAL 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVP---CL 178
PV + P+VC+IE+GGTVGDIES ++EA +Q + R+ C HV+ VP
Sbjct 131 IPVDEDGLE-PQVCVIELGGTVGDIESMPFIEAFRQFQFKVKRENFCNIHVSLVPQPSST 189
Query 179 GEQKTK 184
GEQKTK
Sbjct 190 GEQKTK 195
> 7294316
Length=627
Score = 226 bits (576), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 107/186 (57%), Positives = 138/186 (74%), Gaps = 5/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SS G +L+S G+ V++IKIDPY+N DAGT SPYEHGE +VLDDG EVDLD
Sbjct 11 ISGVGKGVIASSFGTLLKSCGLDVTSIKIDPYINIDAGTFSPYEHGEVYVLDDGAEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL V+L R +NIT+GK+Y VI +ER G +LGK VQ++PH+T AIQ+WV RVA
Sbjct 71 LGNYERFLDVTLHRDNNITTGKIYKLVIEKERTGEYLGKTVQVVPHITDAIQEWVERVAQ 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVP---CL 178
PV + P+VC++E+GGT+GDIE ++EA +Q + R+ CLAHV+ VP
Sbjct 131 TPV--QGSSKPQVCIVELGGTIGDIEGMPFVEAFRQFQFRVKRENFCLAHVSLVPLPKAT 188
Query 179 GEQKTK 184
GE KTK
Sbjct 189 GEPKTK 194
> At4g20320
Length=553
Score = 226 bits (575), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 105/175 (60%), Positives = 135/175 (77%), Gaps = 1/175 (0%)
Query 5 LGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLDLGN 64
LGKG T SSIG++L+S G RV+AIKIDPYLN DAGTMSP EHGE +VLDDGGEVDLDLGN
Sbjct 14 LGKGVTASSIGLILKSCGFRVTAIKIDPYLNIDAGTMSPIEHGEVYVLDDGGEVDLDLGN 73
Query 65 YERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAPQPV 124
YERF+ + L+ ++NIT+GKVY V+ +ER+G +LGK VQ++PH+T AIQ+W+ R A PV
Sbjct 74 YERFMDIKLTSENNITTGKVYKHVLEKERRGDYLGKTVQVVPHITDAIQKWIERAARIPV 133
Query 125 LHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCLG 179
+ +VC+IE+GGT+GDIES ++EA+ Q S +G D CL HV+ VP L
Sbjct 134 -DGQSGPADVCVIELGGTIGDIESMPFIEALGQFSYRVGTDNFCLIHVSLVPVLN 187
> SPAC10F6.03c
Length=600
Score = 216 bits (550), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 110/184 (59%), Positives = 145/184 (78%), Gaps = 2/184 (1%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SS G++L++LG++V++IKIDPY+N DAGTMSP EHGE FVL+DGGEVDLD
Sbjct 11 ISGIGKGVIASSTGLLLKTLGLKVTSIKIDPYMNIDAGTMSPLEHGEVFVLNDGGEVDLD 70
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYER+L V+L+ +NIT+GKVYS VI +ER+G +LGK VQI+PHVT IQ WV RVA
Sbjct 71 LGNYERYLNVTLTHDNNITTGKVYSNVIQKERRGDYLGKTVQIVPHVTNEIQDWVERVAR 130
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPCL-GE 180
PV + + P+VC++E+GGTVGDIESA ++EA++Q +G + HV+ VP + GE
Sbjct 131 IPV-DQSGEEPDVCIVELGGTVGDIESAAFVEAMRQFQFRVGHENFVSIHVSLVPVINGE 189
Query 181 QKTK 184
QKTK
Sbjct 190 QKTK 193
> CE19014
Length=599
Score = 215 bits (548), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 105/186 (56%), Positives = 137/186 (73%), Gaps = 4/186 (2%)
Query 2 MSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDLD 61
+SG+GKG SS+GV+L++ G +VSAIKIDPYLN DAGT SPYEHGE FVLDDGGEVDLD
Sbjct 17 ISGVGKGIISSSLGVLLKNNGYKVSAIKIDPYLNEDAGTFSPYEHGEVFVLDDGGEVDLD 76
Query 62 LGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVAP 121
LGNYERFL + L+R +NIT+GK++ V+ +ER+G + GK VQ+IPH+T AI W+ RVA
Sbjct 77 LGNYERFLDIRLTRDNNITTGKMFKHVMERERRGDYCGKTVQMIPHLTDAIVDWIERVAR 136
Query 122 QPVLHTAADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFV---PCL 178
PV T ++ P+VC+IE+GGT+GDIE YL A ++ R ++ L HV+ +
Sbjct 137 IPVDGT-SEQPDVCIIELGGTIGDIEGMTYLSAFERFQRPTLKNDLMNVHVSLILHPNAT 195
Query 179 GEQKTK 184
GE KTK
Sbjct 196 GEPKTK 201
> ECU11g0880
Length=535
Score = 164 bits (415), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 86/189 (45%), Positives = 120/189 (63%), Gaps = 5/189 (2%)
Query 1 TMSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDL 60
+SG+GKG SSIG +L+S G V+ KIDPYLN +AG M PYEHGE +VLDDG E D+
Sbjct 10 VISGVGKGIVSSSIGALLKSRGHVVTHFKIDPYLNYNAGRMHPYEHGEVYVLDDGHECDM 69
Query 61 DLGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVA 120
D GNYERF G+ LS ++I G++ ++ ER+G FLGK +QI PH+ + + + VA
Sbjct 70 DFGNYERFNGIKLSGANSIPGGRLLHDIVKCEREGSFLGKTLQINPHIIDEVIRRIRAVA 129
Query 121 PQPVLHTA---ADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPC 177
PV A P+V ++E+GGTVG+ ES++Y EA+ + +G+ V ++
Sbjct 130 DTPVESFGGGQAAVPDVVVVELGGTVGEYESSIYTEALAKFQYVVGKANCAFVSVDYIVE 189
Query 178 L--GEQKTK 184
L GEQKTK
Sbjct 190 LETGEQKTK 198
> ECU11g0480
Length=535
Score = 164 bits (415), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 86/189 (45%), Positives = 120/189 (63%), Gaps = 5/189 (2%)
Query 1 TMSGLGKGTTISSIGVVLRSLGVRVSAIKIDPYLNSDAGTMSPYEHGECFVLDDGGEVDL 60
+SG+GKG SSIG +L+S G V+ KIDPYLN +AG M PYEHGE +VLDDG E D+
Sbjct 10 VISGVGKGIVSSSIGALLKSRGHVVTHFKIDPYLNYNAGRMHPYEHGEVYVLDDGHECDM 69
Query 61 DLGNYERFLGVSLSRQHNITSGKVYSQVISQERQGHFLGKAVQIIPHVTAAIQQWVLRVA 120
D GNYERF G+ LS ++I G++ ++ ER+G FLGK +QI PH+ + + + VA
Sbjct 70 DFGNYERFNGIKLSGANSIPGGRLLHDIVKCEREGSFLGKTLQINPHIIDEVIRRIRAVA 129
Query 121 PQPVLHTA---ADTPEVCLIEVGGTVGDIESAVYLEAIQQLSRNLGRDKLCLAHVAFVPC 177
PV A P+V ++E+GGTVG+ ES++Y EA+ + +G+ V ++
Sbjct 130 DTPVESFGGGQAAVPDVVVVELGGTVGEYESSIYTEALAKFQYVVGKANCAFVSVDYIVE 189
Query 178 L--GEQKTK 184
L GEQKTK
Sbjct 190 LETGEQKTK 198
> At4g11030
Length=666
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 27/49 (55%), Gaps = 7/49 (14%)
Query 96 HFLGKAVQIIPHVTAAIQQWVLRVAPQPVLHTAADTPEVCLIEVGGTVG 144
HFLG +V A++ + + ++ P+ H E C+I+VGG++G
Sbjct 257 HFLG-------NVNASLSEKDVYISYLPLAHVFDRAIEECIIQVGGSIG 298
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2950576972
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40