bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2189_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
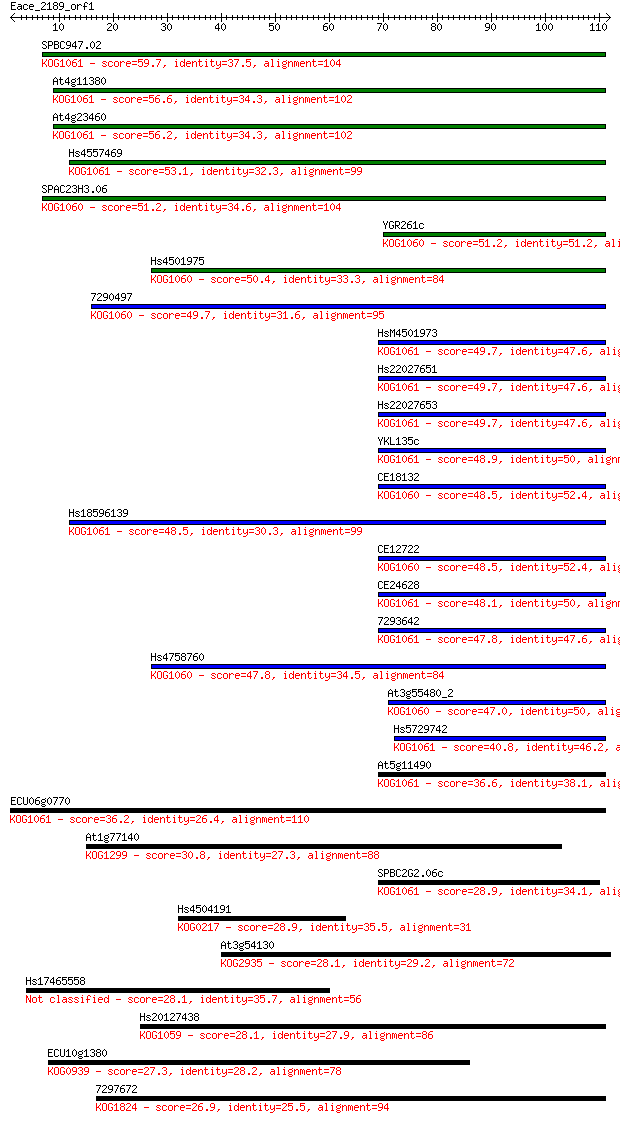
SPBC947.02 59.7 1e-09
At4g11380 56.6 1e-08
At4g23460 56.2 1e-08
Hs4557469 53.1 1e-07
SPAC23H3.06 51.2 5e-07
YGR261c 51.2 5e-07
Hs4501975 50.4 8e-07
7290497 49.7 1e-06
HsM4501973 49.7 1e-06
Hs22027651 49.7 1e-06
Hs22027653 49.7 1e-06
YKL135c 48.9 2e-06
CE18132 48.5 3e-06
Hs18596139 48.5 3e-06
CE12722 48.5 3e-06
CE24628 48.1 4e-06
7293642 47.8 5e-06
Hs4758760 47.8 5e-06
At3g55480_2 47.0 9e-06
Hs5729742 40.8 7e-04
At5g11490 36.6 0.012
ECU06g0770 36.2 0.016
At1g77140 30.8 0.61
SPBC2G2.06c 28.9 2.5
Hs4504191 28.9 2.6
At3g54130 28.1 3.9
Hs17465558 28.1 4.1
Hs20127438 28.1 4.2
ECU10g1380 27.3 6.5
7297672 26.9 8.3
> SPBC947.02
Length=683
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 55/104 (52%), Gaps = 14/104 (13%)
Query 7 KLVSSSESNYFKEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEG 66
KL SS FK+ E S++++ L S ++ ++R IAA G +
Sbjct 4 KLFQSSRFKAFKKSE--TSELQKGLVSQYAYERIDAVKRTIAAMTV----------GKDV 51
Query 67 GSSLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D LKNL+T D LK+LVYLYL+ +A PDL +L+VN
Sbjct 52 SS--LFPDVLKNLATRDITLKKLVYLYLINYAKTHPDLCILAVN 93
> At4g11380
Length=894
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 35/105 (33%), Positives = 57/105 (54%), Gaps = 17/105 (16%)
Query 9 VSSSESNYF---KEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAE 65
+S +S YF K+GEI +++++L+S + + +++ IAA
Sbjct 1 MSGHDSKYFSTTKKGEI--PELKEELNSQYKDKRKDAVKKVIAAMTVGK----------- 47
Query 66 GGSSLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPDLA+L+VN
Sbjct 48 -DVSSLFTDVVNCMQTENLELKKLVYLYLINYAKSQPDLAILAVN 91
> At4g23460
Length=893
Score = 56.2 bits (134), Expect = 1e-08, Method: Composition-based stats.
Identities = 35/105 (33%), Positives = 57/105 (54%), Gaps = 17/105 (16%)
Query 9 VSSSESNYF---KEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAE 65
+S +S YF K+GEI +++++L+S + + +++ IAA
Sbjct 1 MSGHDSKYFSTTKKGEI--PELKEELNSQYKDKRKDAVKKVIAAMTVGK----------- 47
Query 66 GGSSLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPDLA+L+VN
Sbjct 48 -DVSSLFTDVVNCMQTENLELKKLVYLYLINYAKSQPDLAILAVN 91
> Hs4557469
Length=937
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/102 (31%), Positives = 55/102 (53%), Gaps = 17/102 (16%)
Query 12 SESNYF---KEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGGS 68
++S YF K+GEI +++ +L++ E + +++ IAA
Sbjct 2 TDSKYFTTNKKGEI--FELKAELNNEKKEKRKEAVKKVIAAMTVGK------------DV 47
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPD+A+++VN
Sbjct 48 SSLFPDVVNCMQTDNLELKKLVYLYLMNYAKSQPDMAIMAVN 89
> SPAC23H3.06
Length=745
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 58/104 (55%), Gaps = 12/104 (11%)
Query 7 KLVSSSESNYFKEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEG 66
K ++ S S F+E E+ S + + L+S + KL + +A GE
Sbjct 18 KQIAKSSSLSFEENELSHSDLLRLLNSNSDAGKLEAINFILAQMMH-------GE----- 65
Query 67 GSSLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
SL+F D +K +++ + E++RLV++YL+Q+A PDLALLSVN
Sbjct 66 NMSLYFPDVVKLVASENPEIRRLVHIYLLQYAEFNPDLALLSVN 109
> YGR261c
Length=809
Score = 51.2 bits (121), Expect = 5e-07, Method: Composition-based stats.
Identities = 21/41 (51%), Positives = 33/41 (80%), Gaps = 0/41 (0%)
Query 70 LFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
L+FAD +KN++T D ++KRL++LYL++ A P+L LLS+N
Sbjct 81 LYFADVVKNITTNDTKVKRLIHLYLLRFAENDPNLTLLSIN 121
> Hs4501975
Length=1093
Score = 50.4 bits (119), Expect = 8e-07, Method: Composition-based stats.
Identities = 28/84 (33%), Positives = 46/84 (54%), Gaps = 12/84 (14%)
Query 27 IRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGGSSLFFADFLKNLSTTDFEL 86
++Q L S KL ++R + G +S F +KN+++ + E+
Sbjct 47 LKQMLESNKDSAKLDAMKRIV------------GMIAKGKNASELFPAVVKNVASKNIEI 94
Query 87 KRLVYLYLVQHAHEQPDLALLSVN 110
K+LVY+YLV++A EQ DLALLS++
Sbjct 95 KKLVYVYLVRYAEEQQDLALLSIS 118
> 7290497
Length=1160
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 49/95 (51%), Gaps = 12/95 (12%)
Query 16 YFKEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGGSSLFFADF 75
+F+ + ++Q L S KL ++R I G +S F
Sbjct 37 FFQSDGRKHDDLKQMLDSNKDGLKLEAMKRII------------GMIARGRDASDLFPAV 84
Query 76 LKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
+KN+ + + E+K+LVY+YLV++A EQ DLALLS++
Sbjct 85 VKNVVSKNIEVKKLVYVYLVRYAEEQQDLALLSIS 119
> HsM4501973
Length=949
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPD+A+++VN
Sbjct 48 SALFPDVVNCMQTDNLELKKLVYLYLMNYAKSQPDMAIMAVN 89
> Hs22027651
Length=949
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPD+A+++VN
Sbjct 48 SALFPDVVNCMQTDNLELKKLVYLYLMNYAKSQPDMAIMAVN 89
> Hs22027653
Length=942
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPD+A+++VN
Sbjct 48 SALFPDVVNCMQTDNLELKKLVYLYLMNYAKSQPDMAIMAVN 89
> YKL135c
Length=726
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 21/42 (50%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D LKN++T D E K+LVYLY++ +A P+L +L+VN
Sbjct 61 SSLFPDVLKNIATIDVEQKKLVYLYVMNYAETHPELCILAVN 102
> CE18132
Length=896
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 22/42 (52%), Positives = 33/42 (78%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S FA +KN++ + ELK+LV++YLV++A EQ DLALLS++
Sbjct 49 SELFAAVVKNVAAKNVELKKLVFVYLVRYAEEQQDLALLSIS 90
> Hs18596139
Length=863
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 30/102 (29%), Positives = 54/102 (52%), Gaps = 17/102 (16%)
Query 12 SESNYF---KEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGGS 68
++S YF K+ EI +++ +LS+ E + +++ IA
Sbjct 2 TDSKYFTTNKKAEI--FELKAELSNEKKEKRKEAVKKVIAVITVGK------------DV 47
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + +LK+LVYLYL+ +A+ QPD+A+++VN
Sbjct 48 SSVFPDIVNCMQTDNLKLKKLVYLYLMNYANSQPDMAIIAVN 89
> CE12722
Length=906
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 22/42 (52%), Positives = 33/42 (78%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S FA +KN++ + ELK+LV++YLV++A EQ DLALLS++
Sbjct 49 SELFAAVVKNVAAKNVELKKLVFVYLVRYAEEQQDLALLSIS 90
> CE24628
Length=827
Score = 48.1 bits (113), Expect = 4e-06, Method: Composition-based stats.
Identities = 21/42 (50%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPDLA+++VN
Sbjct 48 SALFPDVVNCMQTDNVELKKLVYLYLMNYAKSQPDLAIMAVN 89
> 7293642
Length=921
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F D + + T + ELK+LVYLYL+ +A QPD+A+++VN
Sbjct 48 SALFPDVVNCMQTDNLELKKLVYLYLMNYAKSQPDMAIMAVN 89
> Hs4758760
Length=1081
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 48/84 (57%), Gaps = 12/84 (14%)
Query 27 IRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGGSSLFFADFLKNLSTTDFEL 86
+++ L + KL ++R +A A + S LF A +KN++ + E+
Sbjct 42 LKEMLDTNKDSLKLEAMKRIVAMIARG-----------KNASDLFPA-VVKNVACKNIEV 89
Query 87 KRLVYLYLVQHAHEQPDLALLSVN 110
K+LVY+YLV++A EQ DLALLS++
Sbjct 90 KKLVYVYLVRYAEEQQDLALLSIS 113
> At3g55480_2
Length=1113
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 20/40 (50%), Positives = 32/40 (80%), Gaps = 0/40 (0%)
Query 71 FFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
FF +KN+++ E+K+LVYLYL+Q+A ++P+ ALLS+N
Sbjct 46 FFPQVVKNVASQSSEVKKLVYLYLLQYAEKRPNEALLSIN 85
> Hs5729742
Length=739
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 27/39 (69%), Gaps = 0/39 (0%)
Query 72 FADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
F + +K +T D K+LVYLY+ +A +PDLALL++N
Sbjct 50 FMEMVKASATVDIVQKKLVYLYMCTYAPLKPDLALLAIN 88
> At5g11490
Length=841
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
S F + + +T+D LK++ YLY+ +A PDL+LL++N
Sbjct 62 SSVFGEMVMCSATSDIVLKKMCYLYVGNYAKGNPDLSLLTIN 103
> ECU06g0770
Length=778
Score = 36.2 bits (82), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 47/112 (41%), Gaps = 22/112 (19%)
Query 1 GVRALLKLVSSSESNYFKEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPG 60
G+R L + F + SQI + +SS N + ++ G R ++
Sbjct 11 GIRKLFN------DDSFVANPLGRSQIVKYISSSNIKKQIQGARSLVSR----------- 53
Query 61 EFGAEGGSSLF--FADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
+ G LF D +K T D E KR++ LYLV++ P L+ +N
Sbjct 54 ---MQCGEDLFELCNDVIKATDTRDVEFKRMLNLYLVRYTQGWPAKQLICIN 102
> At1g77140
Length=550
Score = 30.8 bits (68), Expect = 0.61, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 42/91 (46%), Gaps = 10/91 (10%)
Query 15 NYFKEGEIQASQIRQQLSSPNTETKLVGLRRAIA---AEAAAACTYAPGEFGAEGGSSLF 71
NY + ++Q + + + ++ +KLV R+ + E AC G +G +
Sbjct 326 NYPEYKKMQGN-VSKHVTLVTEMSKLVEARKLMTVSQTEQDLACN------GGQGAAYEA 378
Query 72 FADFLKNLSTTDFELKRLVYLYLVQHAHEQP 102
D L N S +D + RLV LY +++ E P
Sbjct 379 VTDLLNNESVSDIDRLRLVMLYALRYEKENP 409
> SPBC2G2.06c
Length=677
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 69 SLFFADFLKNLSTTDFELKRLVYLYLVQHAHEQPDLALLSV 109
S F + ++ + + ELK+L YLYL +A +P A +V
Sbjct 49 SSLFPSVISSMESNNLELKKLCYLYLKIYASVKPTEAKRAV 89
> Hs4504191
Length=1360
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 32 SSPNTETKLVGLRRAIAAEAAAACTYAPGEF 62
S P + GLRR++A A +C ++PG+
Sbjct 65 SPPKAKNLNGGLRRSVAPAAPTSCDFSPGDL 95
> At3g54130
Length=280
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 33/72 (45%), Gaps = 14/72 (19%)
Query 40 LVGLRRAIAAEAAAACTYAPGEFGAEGGSSLFFADFLKNLSTTDFELKRLVYLYLVQHAH 99
L G R + E AA +APG+F AE ++ DF ++ ++Q A
Sbjct 46 LDGKERQVMLEGAAVGGFAPGDFLAEESHNVSLG--------GDFSIQ------VLQKAL 91
Query 100 EQPDLALLSVNC 111
E DL ++ +NC
Sbjct 92 EVWDLQVIPLNC 103
> Hs17465558
Length=1108
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 27/60 (45%), Gaps = 4/60 (6%)
Query 4 ALLKLVSSSESNYFKEGEIQASQIRQQLSSPNTETKLVGLRRAIAAE----AAAACTYAP 59
ALLKL S + + G++Q Q +S + L GL+ A E ACT AP
Sbjct 6 ALLKLQESRKGDVHAGGKVQGEQREGWVSGSTRDVLLPGLQTAQTEEEEDKKPKACTVAP 65
> Hs20127438
Length=1153
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 16/88 (18%)
Query 25 SQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGGSSLFFADF--LKNLSTT 82
+I+Q+L N IA +A A C + G + +A F ++ +S +
Sbjct 41 DEIKQELKQDN-----------IAVKANAVCKLT---YLQMLGYDISWAAFNIIEVMSAS 86
Query 83 DFELKRLVYLYLVQHAHEQPDLALLSVN 110
F KR+ YL Q HE D+ +L+ N
Sbjct 87 KFTFKRIGYLAASQSFHEGTDVIMLTTN 114
> ECU10g1380
Length=2410
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 22/78 (28%), Positives = 32/78 (41%), Gaps = 4/78 (5%)
Query 8 LVSSSESNYFKEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGG 67
LV S +S Y K G + S R SP+ L+ RAI T GEF + G
Sbjct 328 LVESQKSKYLKTGVVMDSLSRFYEYSPDVRYHLL---RAIKMHCRETSTRGIGEFISRNG 384
Query 68 SSLFFADFLKNLSTTDFE 85
+ ++L + D +
Sbjct 385 VRI-LTEYLSHAEKYDLD 401
> 7297672
Length=1248
Score = 26.9 bits (58), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 41/94 (43%), Gaps = 3/94 (3%)
Query 17 FKEGEIQASQIRQQLSSPNTETKLVGLRRAIAAEAAAACTYAPGEFGAEGGSSLFFADFL 76
+ E ++++ IR+ P T GL +AA C Y E G + + F D +
Sbjct 1069 YSETKVKSELIREVEMGPFKHTVDDGLD---IRKAAFECMYTLLEQGLDRVDVMQFLDHV 1125
Query 77 KNLSTTDFELKRLVYLYLVQHAHEQPDLALLSVN 110
+ +++K L YL + A PD LL ++
Sbjct 1126 QAGLCDHYDIKMLTYLMTARLAILCPDKVLLRLD 1159
Lambda K H
0.317 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40