bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2219_orf1
Length=110
Score E
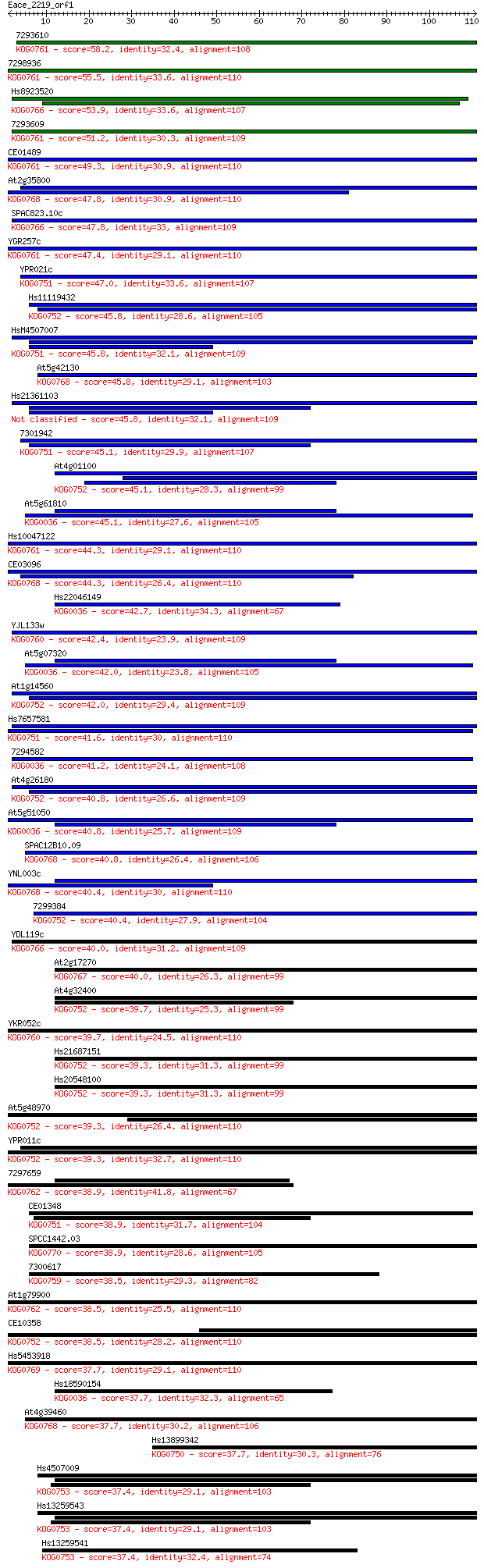
Sequences producing significant alignments: (Bits) Value
7293610 58.2 4e-09
7298936 55.5 2e-08
Hs8923520 53.9 8e-08
7293609 51.2 4e-07
CE01489 49.3 2e-06
At2g35800 47.8 5e-06
SPAC823.10c 47.8 6e-06
YGR257c 47.4 8e-06
YPR021c 47.0 1e-05
Hs11119432 45.8 2e-05
HsM4507007 45.8 2e-05
At5g42130 45.8 2e-05
Hs21361103 45.8 2e-05
7301942 45.1 3e-05
At4g01100 45.1 4e-05
At5g61810 45.1 4e-05
Hs10047122 44.3 6e-05
CE03096 44.3 6e-05
Hs22046149 42.7 2e-04
YJL133w 42.4 2e-04
At5g07320 42.0 2e-04
At1g14560 42.0 3e-04
Hs7657581 41.6 3e-04
7294582 41.2 5e-04
At4g26180 40.8 5e-04
At5g51050 40.8 6e-04
SPAC12B10.09 40.8 6e-04
YNL003c 40.4 8e-04
7299384 40.4 9e-04
YDL119c 40.0 0.001
At2g17270 40.0 0.001
At4g32400 39.7 0.001
YKR052c 39.7 0.002
Hs21687151 39.3 0.002
Hs20548100 39.3 0.002
At5g48970 39.3 0.002
YPR011c 39.3 0.002
7297659 38.9 0.002
CE01348 38.9 0.002
SPCC1442.03 38.9 0.003
7300617 38.5 0.003
At1g79900 38.5 0.003
CE10358 38.5 0.003
Hs5453918 37.7 0.005
Hs18590154 37.7 0.006
At4g39460 37.7 0.006
Hs13899342 37.7 0.006
Hs4507009 37.4 0.006
Hs13259543 37.4 0.006
Hs13259541 37.4 0.007
> 7293610
Length=413
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 58/109 (53%), Gaps = 8/109 (7%)
Query 3 RGAAVAIFNPISIVKTRMESS-LGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSG 61
R AV +P+ +++T+M+S + M Q+++++ L +RG TI RD PFSG
Sbjct 192 RILAVTCVSPVELIRTKMQSQRMTHAEMFGTIRQVVQSQGVLGLWRGLPPTILRDVPFSG 251
Query 62 LFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+++ Y LKS GV + +F GA++ + A+ IT PFD
Sbjct 252 IYWTCYEYLKSSFGVVEPTFSF-------SFAAGAISGSVAATITTPFD 293
> 7298936
Length=449
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 60/114 (52%), Gaps = 13/114 (11%)
Query 1 VARGAAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLD--W--FRGTFATICRD 56
AR AV + +PI +V+T+M++ + LQ +R+ L W +RG TI RD
Sbjct 243 TARICAVTVVSPIELVRTKMQAQRQTYAQ---MLQFVRSVVALQGVWGLWRGLRPTILRD 299
Query 57 FPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
PFSG+++ +Y +LK ++G + +S F G +A A+ +T PFD
Sbjct 300 VPFSGIYWPIYESLKQNLGHGSQPSFSLS------FLAGVMAGTVAAIVTTPFD 347
> Hs8923520
Length=304
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 53/111 (47%), Gaps = 10/111 (9%)
Query 2 ARGAAVAIFNPISIVKTRMES-SLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
+R A +PI+++KTR ES G + A + +E F G AT+ RD PFS
Sbjct 133 SRSVAGVCMSPITVIKTRYESGKYGYESIYAALRSIYHSEGHRGLFSGLTATLLRDAPFS 192
Query 61 GLFFVVY---VNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHP 108
G++ + Y N+ H V+A + + NF CG A AS +T P
Sbjct 193 GIYLMFYNQTKNIVPHDQVDA------TLIPITNFSCGIFAGILASLVTQP 237
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 49/106 (46%), Gaps = 12/106 (11%)
Query 9 IFNPISIVKTRMESSLGSG------GMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGL 62
+F P+ ++KTR+++ S GM L+++R E L ++G +I R P G+
Sbjct 44 LFQPLDLLKTRLQTLQPSDHGSRRVGMLAVLLKVVRTESLLGLWKGMSPSIVRCVPGVGI 103
Query 63 FFVVYVNLKSHM--GVEADSRERVSCYALQNFGCGAVAAATASAIT 106
+F +LK + G + E V + G +VA S IT
Sbjct 104 YFGTLYSLKQYFLRGHPPTALESV----MLGVGSRSVAGVCMSPIT 145
> 7293609
Length=443
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 8/110 (7%)
Query 2 ARGAAVAIFNPISIVKTRMESS-LGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
+R V PI +V+ +M+S + + + L+R L +RG T+ RD PFS
Sbjct 229 SRTIVVTAITPIEMVRIKMQSEYMTYAELWRVLRSLIRQHGILGLWRGWPPTVMRDAPFS 288
Query 61 GLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
G ++ VY +K V + L +F GA++ A A+ +T PFD
Sbjct 289 GTYWAVYEAIKRAFSVTEPT-------FLFSFLTGAISGAVATFVTMPFD 331
> CE01489
Length=328
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 55/115 (47%), Gaps = 12/115 (10%)
Query 1 VARGAAVAIFNPISIVKTRMESS-LGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPF 59
VAR AV + +PI +++T+M+S L + + + ++ G T+ RD PF
Sbjct 151 VARTIAVTVVSPIEMIRTKMQSKRLTYHEIGHLVRSSMATKGISSFYLGWTPTMLRDIPF 210
Query 60 SGLFFVVY----VNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
SG+++ Y NL+ G + + + +F GA A AS THPFD
Sbjct 211 SGIYWAGYDLFKTNLQRRQGPDHNP-------FVVSFVSGAAAGVVASIFTHPFD 258
> At2g35800
Length=844
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 55/107 (51%), Gaps = 7/107 (6%)
Query 4 GAAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLF 63
G AV I P ++K R+++ + + + +A + + + P +FRGT AT+CR+ P +
Sbjct 665 GTAVRI--PCEVLKQRLQAGMFNN-VGEAIVGTWKQDGPSGFFRGTGATLCREVPLYVVG 721
Query 64 FVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+Y K M +A RE A + GAV+ A+ +T PFD
Sbjct 722 MGLYAESK-KMVAQALGRE---LEAWETIAVGAVSGGIAAVVTTPFD 764
Score = 35.0 bits (79), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 35/82 (42%), Gaps = 2/82 (2%)
Query 1 VARGAAVAIFNPISIVKTRMESSLGSG--GMSQAFLQLLRNERPLDWFRGTFATICRDFP 58
V+ G A + P ++KTRM ++ MS + +LRNE PL F+G P
Sbjct 751 VSGGIAAVVTTPFDVMKTRMMTATPGRPISMSMVVVSILRNEGPLGLFKGAVPRFFWVAP 810
Query 59 FSGLFFVVYVNLKSHMGVEADS 80
+ F Y K M D+
Sbjct 811 LGAMNFAGYELAKKAMQKNEDA 832
> SPAC823.10c
Length=296
Score = 47.8 bits (112), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 36/117 (30%), Positives = 56/117 (47%), Gaps = 8/117 (6%)
Query 2 ARGAAVAIFNPISIVKTRMESSLGS-GGMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
AR A F P++++K R ES+L S + + + + E +FRG T RD P +
Sbjct 116 ARAAVGFAFMPVTVIKVRYESNLYSYTTIYSSIRDIWKKEGISGFFRGFGVTALRDAPHA 175
Query 61 GLFFVVYVNLKSHMGVEADS-------RERVSCYALQNFGCGAVAAATASAITHPFD 110
GL+ Y K ++ D + V + N G ++ ATA+AIT+PFD
Sbjct 176 GLYVYFYELSKQNLHKLFDRFSPSSSVQGTVPHRNIVNVMSGLISGATATAITNPFD 232
> YGR257c
Length=366
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 58/124 (46%), Gaps = 14/124 (11%)
Query 1 VARGAAVAIFNPISIVKTRMES------SLGSGGMSQAFLQLLRNERPL-----DWFRGT 49
+AR A P+ +VKT+++S S + M + L R E + F+G
Sbjct 167 IARVFAATSIAPLELVKTKLQSIPRSSKSTKTWMMVKDLLNETRQEMKMVGPSRALFKGL 226
Query 50 FATICRDFPFSGLFFVVYVNLKSHMGVEAD---SRERVSCYALQNFGCGAVAAATASAIT 106
T+ RD PFS +++ Y K + +++ S++ + + +F G ++ A+ T
Sbjct 227 EITLWRDVPFSAIYWSSYELCKERLWLDSTRFASKDANWVHFINSFASGCISGMIAAICT 286
Query 107 HPFD 110
HPFD
Sbjct 287 HPFD 290
> YPR021c
Length=902
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 36/117 (30%), Positives = 55/117 (47%), Gaps = 12/117 (10%)
Query 4 GAAVAIF-NPISIVKTRME--SSLGSGGMSQA---FLQLLRNERPLDWFRGTFATICRDF 57
GA IF NP+ IVK R++ S + QA Q+++ + G A + RD
Sbjct 635 GACQVIFTNPLEIVKIRLQVQSDYVGENIQQANETATQIVKKLGLRGLYNGVAACLMRDV 694
Query 58 PFSGLFFVVYVNLKSHM----GVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
PFS ++F Y +LK + + R R+ + L GA+A A+ +T PFD
Sbjct 695 PFSAIYFPTYAHLKKDLFDFDPNDKTKRNRLKTWEL--LTAGAIAGMPAAFLTTPFD 749
> Hs11119432
Length=320
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 51/108 (47%), Gaps = 4/108 (3%)
Query 6 AVAIFNPISIVKTRMESSLGS---GGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGL 62
A +P+ +++TR + + A + R+E P +++G T+ FP++GL
Sbjct 132 ATLTVHPVDVLRTRFAAQGEPKVYNTLRHAVGTMYRSEGPQVFYKGLAPTLIAIFPYAGL 191
Query 63 FFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
F Y +LK H+ A E LQN CG+ A + +T+P D
Sbjct 192 QFSCYSSLK-HLYKWAIPAEGKKNENLQNLLCGSGAGVISKTLTYPLD 238
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 51/115 (44%), Gaps = 17/115 (14%)
Query 8 AIFNPISIVKTRME----------SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDF 57
A+ +P ++K R + S G+ QA Q+L+ E P +++G
Sbjct 31 ALISPFDVIKIRFQLQHERLSRSDPSAKYHGILQASRQILQEEGPTAFWKGHVPAQILSI 90
Query 58 PFSGLFFVVYVNLKS--HMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+ + F+ + L H G D+RE +++ +F CG +AA A+ HP D
Sbjct 91 GYGAVQFLSFEMLTELVHRGSVYDARE----FSV-HFVCGGLAACMATLTVHPVD 140
> HsM4507007
Length=678
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 53/111 (47%), Gaps = 8/111 (7%)
Query 2 ARGAAVAIFNPISIVKTRME--SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPF 59
A G+ V NP+ IVK R++ + +G A L +LR+ ++G A RD PF
Sbjct 436 AGGSQVIFTNPLEIVKIRLQVAGEITTGPRVSA-LNVLRDLGIFGLYKGAKACFLRDIPF 494
Query 60 SGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
S ++F VY + K + AD V L GA+A A+++ P D
Sbjct 495 SAIYFPVYAHCKLLL---ADENGHVG--GLNLLAAGAMAGVPAASLVTPAD 540
Score = 34.3 bits (77), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 20/123 (16%)
Query 6 AVAIFNPISIVKTRMESSLGSG-----GMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
A ++ P ++KTR++ + +G G+ F ++LR E P +++GT A + R P
Sbjct 532 AASLVTPADVIKTRLQVAARAGQTTYSGVIDCFRKILREEGPSAFWKGTAARVFRSSPQF 591
Query 61 GL-----------FFVVYVNLKSHMGVEADSRERVSCYALQN---FGCGAVAAATASAIT 106
G+ F++ + LK G E + R++ N G +A AT + I
Sbjct 592 GVTLAHYEVLQRWFYIDFGGLKP-AGSEPTPKSRIADLPPANPDHIGGYRLATATFAGIE 650
Query 107 HPF 109
+ F
Sbjct 651 NKF 653
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 10/52 (19%)
Query 6 AVAIFNPISIVKTRMESSLGSGGMS---------QAFLQLLRNERPLDWFRG 48
A A++ PI +VKTRM++ GSG + F ++LR E +RG
Sbjct 341 ATAVY-PIDLVKTRMQNQRGSGSVVGELMYKNSFDCFKKVLRYEGFFGLYRG 391
> At5g42130
Length=412
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/103 (29%), Positives = 52/103 (50%), Gaps = 4/103 (3%)
Query 8 AIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVY 67
AI P ++ RM++ SG Q L++L + L + G AT+ R+ P L + +
Sbjct 224 AIMVPKELITQRMQAG-ASGRSYQVLLKILEKDGILGLYAGYSATLLRNLPAGVLSYSSF 282
Query 68 VNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
LK+ + + ++ LQ+ CGA+A A +++IT P D
Sbjct 283 EYLKAAV---LEKTKQSHLEPLQSVCCGALAGAISASITTPLD 322
> Hs21361103
Length=678
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 53/111 (47%), Gaps = 8/111 (7%)
Query 2 ARGAAVAIFNPISIVKTRME--SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPF 59
A G+ V NP+ IVK R++ + +G A L +LR+ ++G A RD PF
Sbjct 436 AGGSQVIFTNPLEIVKIRLQVAGEITTGPRVSA-LNVLRDLGIFGLYKGAKACFLRDIPF 494
Query 60 SGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
S ++F VY + K + AD V L GA+A A+++ P D
Sbjct 495 SAIYFPVYAHCKLLL---ADENGHVG--GLNLLAAGAMAGVPAASLVTPAD 540
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query 6 AVAIFNPISIVKTRMESSLGSG-----GMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
A ++ P ++KTR++ + +G G+ F ++LR E P +++GT A + R P
Sbjct 532 AASLVTPADVIKTRLQVAARAGQTTYSGVIDCFRKILREEGPSAFWKGTAARVFRSSPQF 591
Query 61 GLFFVVYVNLK 71
G+ V Y L+
Sbjct 592 GVTLVTYELLQ 602
Score = 27.3 bits (59), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 10/52 (19%)
Query 6 AVAIFNPISIVKTRMESSLGSGGMS---------QAFLQLLRNERPLDWFRG 48
A A++ PI +VKTRM++ GSG + F ++LR E +RG
Sbjct 341 ATAVY-PIDLVKTRMQNQRGSGSVVGELMYKNSFDCFKKVLRYEGFFGLYRG 391
> 7301942
Length=695
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 55/110 (50%), Gaps = 9/110 (8%)
Query 4 GAAVAIF-NPISIVKTRME--SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
GA+ +F NP+ IVK R++ + SG +A+ ++R ++G A + RD PFS
Sbjct 452 GASQVVFTNPLEIVKIRLQVAGEIASGSKIRAW-SVVRELGLFGLYKGARACLLRDVPFS 510
Query 61 GLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
++F Y + K+ M + D + L GA+A A+++ P D
Sbjct 511 AIYFPTYAHTKAMMA-DKDGYN----HPLTLLAAGAIAGVPAASLVTPAD 555
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 36/71 (50%), Gaps = 5/71 (7%)
Query 6 AVAIFNPISIVKTRMESSLGSG-----GMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
A ++ P ++KTR++ SG G+ A +++ E P +++GT A + R P
Sbjct 547 AASLVTPADVIKTRLQVVARSGQTTYTGVWDATKKIMAEEGPRAFWKGTAARVFRSSPQF 606
Query 61 GLFFVVYVNLK 71
G+ V Y L+
Sbjct 607 GVTLVTYELLQ 617
> At4g01100
Length=352
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 45/106 (42%), Gaps = 7/106 (6%)
Query 12 PISIVKTRMESSLGSG-----GMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVV 66
P+ +V+ R+ + G++ A +LR E P +RG ++ P+ GL F V
Sbjct 161 PMDMVRGRLTVQTANSPYQYRGIAHALATVLREEGPRALYRGWLPSVIGVVPYVGLNFSV 220
Query 67 YVNLKSHMGVEADSR--ERVSCYALQNFGCGAVAAATASAITHPFD 110
Y +LK + E E + CGA+A I +P D
Sbjct 221 YESLKDWLVKENPYGLVENNELTVVTRLTCGAIAGTVGQTIAYPLD 266
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 30/86 (34%), Gaps = 3/86 (3%)
Query 28 GMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNLKS---HMGVEADSRERV 84
G Q + R E F+G R P S + F Y + +M + E
Sbjct 78 GTVQGLKHIWRTEGLRGLFKGNGTNCARIVPNSAVKFFSYEQASNGILYMYRQRTGNENA 137
Query 85 SCYALQNFGCGAVAAATASAITHPFD 110
L G GA A A + T+P D
Sbjct 138 QLTPLLRLGAGATAGIIAMSATYPMD 163
Score = 28.9 bits (63), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 19 RMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNLKSHMGVE 77
R +SL GM AF + +R+E ++G + P + FV Y +K +GVE
Sbjct 289 RSTASLEYTGMVDAFRKTVRHEGFGALYKGLVPNSVKVVPSIAIAFVTYEMVKDVLGVE 347
> At5g61810
Length=476
Score = 45.1 bits (105), Expect = 4e-05, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 12 PISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNLK 71
P+ +++TRM++ M Q FL+ LR E ++RG F + P + + ++VY +K
Sbjct 411 PLQVIRTRMQADSSKTSMGQEFLKTLRGEGLKGFYRGIFPNFFKVIPSASISYLVYEAMK 470
Query 72 SHMGVE 77
++ ++
Sbjct 471 KNLALD 476
Score = 33.5 bits (75), Expect = 0.095, Method: Composition-based stats.
Identities = 27/111 (24%), Positives = 49/111 (44%), Gaps = 13/111 (11%)
Query 5 AAVAIFNPISIVKTRME---SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSG 61
A AI+ P+ +VKTR++ S +G+ + + + E P ++RG ++ P++G
Sbjct 309 AQTAIY-PMDLVKTRLQTFVSEVGTPKLWKLTKDIWIQEGPRAFYRGLCPSLIGIIPYAG 367
Query 62 LFFVVYVNLKSHMGVEADSRERV---SCYALQNFGCGAVAAATASAITHPF 109
+ Y LK SR + L GCG + A ++ +P
Sbjct 368 IDLAAYETLKDL------SRAHFLHDTAGPLIQLGCGMTSGALGASCVYPL 412
> Hs10047122
Length=338
Score = 44.3 bits (103), Expect = 6e-05, Method: Composition-based stats.
Identities = 32/111 (28%), Positives = 56/111 (50%), Gaps = 4/111 (3%)
Query 1 VARGAAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERP-LDWFRGTFATICRDFPF 59
VAR AV + + + +++T+M+S S F+ +E + +RG T+ RD PF
Sbjct 150 VARFGAVTVISQLELIRTKMQSKKFSYVELHRFVSKKVSEDGWISLWRGWAPTVLRDVPF 209
Query 60 SGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
S +++ Y LK + ++ E + NF GA++ + A+ T PFD
Sbjct 210 SAMYWYNYEILKKWLCEKSGLYEPT---FMINFTSGALSGSFAAVATLPFD 257
> CE03096
Length=269
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/110 (26%), Positives = 55/110 (50%), Gaps = 4/110 (3%)
Query 1 VARGAAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
+A AA A+ P + K R + + ++ +++ + ++RG +T+ R+ PFS
Sbjct 98 LAEIAACAVRVPTELCKQRGQVNKNER-LTLICKEIMETKGIRGFYRGYGSTVAREIPFS 156
Query 61 GLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+ F ++ LK + A+ +E C L+ CG+VA A+ +T P D
Sbjct 157 IIQFPIWEALKRAV---ANKKESGRCSPLEGAACGSVAGFIAAGLTTPLD 203
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 34/82 (41%), Gaps = 17/82 (20%)
Query 4 GAAVAI-FNPISIVKTRMESSLG---SGGMSQAFLQLLRNERPLDWFRGTFATICRDFPF 59
G AV I P+ +K+RM+S G +GG D +RG + + P
Sbjct 19 GLAVDIGLYPLDTIKSRMQSKQGFIAAGGFK-------------DIYRGMISVLVGSAPG 65
Query 60 SGLFFVVYVNLKSHMGVEADSR 81
+ +FF+ Y + M + R
Sbjct 66 AAIFFLTYKYINGQMKQVIEER 87
> Hs22046149
Length=469
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 37/72 (51%), Gaps = 5/72 (6%)
Query 12 PISIVKTRMESSLGSGG-----MSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVV 66
P+++V+TRM++ G MS F +LR E +RG + P + +VV
Sbjct 397 PLALVRTRMQAQASIEGAPEVTMSSLFKHILRTEGAFGLYRGLAPNFMKVIPAVSISYVV 456
Query 67 YVNLKSHMGVEA 78
Y NLK +GV++
Sbjct 457 YENLKITLGVQS 468
> YJL133w
Length=314
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 51/109 (46%), Gaps = 7/109 (6%)
Query 2 ARGAAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSG 61
A A+ A+ NP +K R++ + S + Q Q+ ++E ++ T+ + PF+
Sbjct 140 ATTASDALMNPFDTIKQRIQLNT-SASVWQTTKQIYQSEGLAAFYYSYPTTLVMNIPFAA 198
Query 62 LFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
FV+Y + + + + C CG+++ +T +AIT P D
Sbjct 199 FNFVIYESSTKFLNPSNEYNPLIHCL------CGSISGSTCAAITTPLD 241
> At5g07320
Length=479
Score = 42.0 bits (97), Expect = 2e-04, Method: Composition-based stats.
Identities = 16/66 (24%), Positives = 36/66 (54%), Gaps = 0/66 (0%)
Query 12 PISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNLK 71
P+ +V+TRM++ M Q F+ ++ E ++RG + + P + + ++VY +K
Sbjct 414 PLQVVRTRMQADSSKTTMKQEFMNTMKGEGLRGFYRGLLPNLLKVVPAASITYIVYEAMK 473
Query 72 SHMGVE 77
+M ++
Sbjct 474 KNMALD 479
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 27/111 (24%), Positives = 52/111 (46%), Gaps = 11/111 (9%)
Query 5 AAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRN----ERPLDWFRGTFATICRDFPFS 60
A AI+ P+ +VKTR+++ + GG + +L ++ E P +++G F ++ P++
Sbjct 310 AQTAIY-PMDLVKTRLQTCVSEGGKAPKLWKLTKDIWVREGPRAFYKGLFPSLLGIVPYA 368
Query 61 GLFFVVYVNLK--SHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPF 109
G+ Y LK S + D+ L CG + A ++ +P
Sbjct 369 GIDLAAYETLKDLSRTYILQDTEPG----PLIQLSCGMTSGALGASCVYPL 415
> At1g14560
Length=331
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 32/127 (25%), Positives = 51/127 (40%), Gaps = 24/127 (18%)
Query 2 ARGAAVAIFNPISIVKTRM-------ESSLGSG-----------GMSQAFLQLLRNERPL 43
A G AV P+ + +T++ SL G G+ + + P
Sbjct 129 AGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYSGIKEVLAMAYKEGGPR 188
Query 44 DWFRGTFATICRDFPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATAS 103
+RG T+ P++GL F +Y LK H+ E + R+ + CGA+A
Sbjct 189 GLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRM------HLPCGALAGLFGQ 242
Query 104 AITHPFD 110
IT+P D
Sbjct 243 TITYPLD 249
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 24/105 (22%), Positives = 50/105 (47%), Gaps = 3/105 (2%)
Query 6 AVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFV 65
AVA I I+ + + G+SQ+ ++L+ + PL +++G A++ R P++ L ++
Sbjct 40 AVAPLERIKILLQTRTNDFKTLGVSQSLKKVLQFDGPLGFYKGNGASVIRIIPYAALHYM 99
Query 66 VYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
Y + + + + + + G+ A TA T+P D
Sbjct 100 TYEVYRDWI---LEKNLPLGSGPIVDLVAGSAAGGTAVLCTYPLD 141
> Hs7657581
Length=675
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 56/111 (50%), Gaps = 8/111 (7%)
Query 2 ARGAAVAIFNPISIVKTRME--SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPF 59
A G+ V NP+ IVK R++ + +G A L ++R+ ++G A RD PF
Sbjct 438 AGGSQVIFTNPLEIVKIRLQVAGEITTGPRVSA-LSVVRDLGFFGIYKGAKACFLRDIPF 496
Query 60 SGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
S ++F Y ++K+ A+ +VS +L GA+A A+++ P D
Sbjct 497 SAIYFPCYAHVKASF---ANEDGQVSPGSL--LLAGAIAGMPAASLVTPAD 542
Score = 34.7 bits (78), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 31/126 (24%), Positives = 56/126 (44%), Gaps = 17/126 (13%)
Query 1 VARGAAVAIFNPISIVKTRMESSLGSG-----GMSQAFLQLLRNERPLDWFRGTFATICR 55
+A A ++ P ++KTR++ + +G G+ F ++LR E P ++G A + R
Sbjct 529 IAGMPAASLVTPADVIKTRLQVAARAGQTTYSGVIDCFRKILREEGPKALWKGAGARVFR 588
Query 56 DFPFSGLFFVVYVNLK----------SHMGVEADSRERVSCYALQ--NFGCGAVAAATAS 103
P G+ + Y L+ MG E + R++ A + G +A AT +
Sbjct 589 SSPQFGVTLLTYELLQRWFYIDFGGVKPMGSEPVPKSRINLPAPNPDHVGGYKLAVATFA 648
Query 104 AITHPF 109
I + F
Sbjct 649 GIENKF 654
> 7294582
Length=370
Score = 41.2 bits (95), Expect = 5e-04, Method: Composition-based stats.
Identities = 26/110 (23%), Positives = 50/110 (45%), Gaps = 4/110 (3%)
Query 2 ARGAAVAIFNPISIVKTRM--ESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPF 59
A G + I P+ ++KTR+ + G++ A +++ + E ++RG I P+
Sbjct 175 AGGISQTIIYPMEVLKTRLALRRTGQYAGIADAAVKIYKQEGVRSFYRGYVPNILGILPY 234
Query 60 SGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPF 109
+G+ VY LK D+ E+ S L CG+ ++ ++P
Sbjct 235 AGIDLAVYETLKRRYIANHDNNEQPSFLVL--LACGSTSSTLGQLCSYPL 282
> At4g26180
Length=325
Score = 40.8 bits (94), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 50/119 (42%), Gaps = 16/119 (13%)
Query 2 ARGAAVAIFNPISIVKTRME----------SSLGSGGMSQAFLQLLRNERPLDWFRGTFA 51
A G AV P+ +V+T++ + G+ F + R +RG
Sbjct 123 AGGTAVLFTYPLDLVRTKLAYQTQVKAIPVEQIIYRGIVDCFSRTYRESGARGLYRGVAP 182
Query 52 TICRDFPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
++ FP++GL F Y +K H V + ++ +S CG+VA +T+P D
Sbjct 183 SLYGIFPYAGLKFYFYEEMKRH--VPPEHKQDISL----KLVCGSVAGLLGQTLTYPLD 235
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/107 (22%), Positives = 45/107 (42%), Gaps = 7/107 (6%)
Query 6 AVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFV 65
AVA I I+ G+ + ++ + E + ++RG A++ R P++ L ++
Sbjct 34 AVAPLERIKILFQTRRDEFKRIGLVGSINKIGKTEGLMGFYRGNGASVARIVPYAALHYM 93
Query 66 VYVNLKSHM--GVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
Y + + G +R L + G+ A TA T+P D
Sbjct 94 AYEEYRRWIIFGFPDTTRG-----PLLDLVAGSFAGGTAVLFTYPLD 135
> At5g51050
Length=487
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 28/116 (24%), Positives = 52/116 (44%), Gaps = 11/116 (9%)
Query 1 VARGAAVAIFNPISIVKTRMESSLGSGGMSQAFL-----QLLRNERPLDWFRGTFATICR 55
+A A A P+ +VKTR+++ G++ L +L +E P +++G F ++
Sbjct 312 MAGAVAQASIYPLDLVKTRLQTYTSQAGVAVPRLGTLTKDILVHEGPRAFYKGLFPSLLG 371
Query 56 DFPFSGLFFVVYVNLK--SHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPF 109
P++G+ Y LK S + D+ L GCG ++ A + +P
Sbjct 372 IIPYAGIDLAAYETLKDLSRTYILQDAEPG----PLVQLGCGTISGALGATCVYPL 423
Score = 32.7 bits (73), Expect = 0.17, Method: Composition-based stats.
Identities = 14/66 (21%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 12 PISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNLK 71
P+ +V+TRM++ MS F + + E ++G + + P + + ++VY +K
Sbjct 422 PLQVVRTRMQAERARTSMSGVFRRTISEEGYRALYKGLLPNLLKVVPAASITYMVYEAMK 481
Query 72 SHMGVE 77
+ ++
Sbjct 482 KSLELD 487
> SPAC12B10.09
Length=345
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 54/107 (50%), Gaps = 4/107 (3%)
Query 5 AAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLR-NERPLDWFRGTFATICRDFPFSGLF 63
AA + P ++K R ++S G+ +L+ N D++ G TI R+ PF+ +
Sbjct 177 AACIVRVPTEVIKQRAQASGGTLSSRNILQTILKSNNVWRDFYAGYGITIAREIPFTLIQ 236
Query 64 FVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
F ++ +LK ++ + ++ A + G++A A+A+T PFD
Sbjct 237 FPIWEHLKLKWRIKHSRNKNLAHEAAIS---GSIAGGIAAALTTPFD 280
> YNL003c
Length=284
Score = 40.4 bits (93), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 49/103 (47%), Gaps = 8/103 (7%)
Query 12 PISIVKTRMESSLGSGGMSQAFLQLLRNERP----LDWFRGTFATICRDFPFSGLFFVVY 67
P +VK R + + Q +LRN+ + +RG TI R+ PF+ + F +Y
Sbjct 114 PAEVVKQRTQVH-STNSSWQTLQSILRNDNKEGLRKNLYRGWSTTIMREIPFTCIQFPLY 172
Query 68 VNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
LK +A+ + +V + + CG++A A+A T P D
Sbjct 173 EYLKKTW-AKANGQSQVEPW--KGAICGSIAGGIAAATTTPLD 212
Score = 27.3 bits (59), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 13/48 (27%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 1 VARGAAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRG 48
+A G A A P+ +KTR+ + + + +++ R E P +F G
Sbjct 199 IAGGIAAATTTPLDFLKTRLMLNKTTASLGSVIIRIYREEGPAVFFSG 246
> 7299384
Length=332
Score = 40.4 bits (93), Expect = 9e-04, Method: Composition-based stats.
Identities = 29/110 (26%), Positives = 52/110 (47%), Gaps = 9/110 (8%)
Query 7 VAIFNPISIVKTRM---ESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLF 63
V I P+ +++TR+ ++S G ++A ++R E P +RG + + + P G
Sbjct 153 VIISTPLDVIRTRLIAQDTSKGYRNATRAVSAIVRQEGPRGMYRGLSSALLQITPLMGTN 212
Query 64 FVVYVNLKSHMG---VEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
F+ Y L S +E R ++ + L G GA + + I +PFD
Sbjct 213 FMAY-RLFSDWACAFLEVSDRSQLPTWTL--LGLGASSGMLSKTIVYPFD 259
> YDL119c
Length=307
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 54/119 (45%), Gaps = 10/119 (8%)
Query 2 ARGAAVAIFNPISIVKTRMESSL-GSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
ARG I PI+++K R ES+L + +A + E +FRG AT RD P++
Sbjct 127 ARGLVGYITMPITVIKVRYESTLYNYSSLKEAITHIYTKEGLFGFFRGFGATCLRDAPYA 186
Query 61 GLFFVVYVNLKSHMGVEADSR---------ERVSCYALQNFGCGAVAAATASAITHPFD 110
GL+ ++Y K + + SR N ++A+ A+ +T PFD
Sbjct 187 GLYVLLYEKSKQLLPMVLPSRFIHYNPEGGFTTYTSTTVNTTSAVLSASLATTVTAPFD 245
> At2g17270
Length=309
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 5/104 (4%)
Query 12 PISIVKTRMESS-LGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNL 70
P +K R+++ + + G+ F ++ R+E + RG F CR+ PFS + F +
Sbjct 131 PFEAIKVRVQTQPMFAKGLLDGFPRVYRSEGLAGFHRGLFPLWCRNLPFSMVMFSTFEQS 190
Query 71 KSHMGVEADSRERVSCYALQNFGCGAVAAATASA----ITHPFD 110
+ + + + C Q G +A TA A I++P D
Sbjct 191 VEFIYQKIIQKRKQDCSKAQQLGVTCLAGYTAGAVGTIISNPAD 234
> At4g32400
Length=392
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 50/100 (50%), Gaps = 4/100 (4%)
Query 12 PISIVKTRMESSLGS-GGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNL 70
P+ +VKTR+ G G+ AFL+++R E P + +RG ++ P++ + Y +L
Sbjct 224 PLELVKTRLTIQRGVYKGIFDAFLKIIREEGPTELYRGLAPSLIGVVPYAATNYFAYDSL 283
Query 71 KSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+ +E++ ++ G++A A +S T P +
Sbjct 284 RKAY-RSFSKQEKIG--NIETLLIGSLAGALSSTATFPLE 320
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/61 (24%), Positives = 29/61 (47%), Gaps = 5/61 (8%)
Query 12 PISIVKTRMESSLGSG-----GMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVV 66
P+ + + M+ SG M A + +L +E L W++G + + P +G+ F+
Sbjct 318 PLEVARKHMQVGAVSGRVVYKNMLHALVTILEHEGILGWYKGLGPSCLKLVPAAGISFMC 377
Query 67 Y 67
Y
Sbjct 378 Y 378
> YKR052c
Length=304
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 52/112 (46%), Gaps = 11/112 (9%)
Query 1 VARGAAVAIFNPISIVKTRME--SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFP 58
+A AA A+ NP VK R++ ++L +++ Q+ +NE ++ T+ + P
Sbjct 129 IATIAADALMNPFDTVKQRLQLDTNLRVWNVTK---QIYQNEGFAAFYYSYPTTLAMNIP 185
Query 59 FSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
F+ F++Y + + + C CG ++ AT +A+T P D
Sbjct 186 FAAFNFMIYESASKFFNPQNSYNPLIHCL------CGGISGATCAALTTPLD 231
> Hs21687151
Length=341
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 50/103 (48%), Gaps = 9/103 (8%)
Query 12 PISIVKTR--MESSLGSG--GMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVY 67
P ++KTR M++ L G+ AF + + E L +RG T+ PFS +VY
Sbjct 126 PTDLIKTRLIMQNILEPSYRGLLHAFSTIYQQEGFLALYRGVSLTVVGALPFSAGSLLVY 185
Query 68 VNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+NL+ + R++ S QNF +AAA ++ PF+
Sbjct 186 MNLEK---IWNGPRDQFSLP--QNFANVCLAAAVTQTLSFPFE 223
> Hs20548100
Length=341
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 50/103 (48%), Gaps = 9/103 (8%)
Query 12 PISIVKTR--MESSLGSG--GMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVY 67
P ++KTR M++ L G+ AF + + E L +RG T+ PFS +VY
Sbjct 126 PTDLIKTRLIMQNILEPSYRGLLHAFSTIYQQEGFLALYRGVSLTVVGALPFSAGSLLVY 185
Query 68 VNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+NL+ + R++ S QNF +AAA ++ PF+
Sbjct 186 MNLEK---IWNGPRDQFSLP--QNFANVCLAAAVTQTLSFPFE 223
> At5g48970
Length=339
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/127 (22%), Positives = 54/127 (42%), Gaps = 17/127 (13%)
Query 1 VARGAAVAIFNPISIVKTRMESSLG------------SG-----GMSQAFLQLLRNERPL 43
++ G + ++ +P+ ++K R + L SG GM QA + R E
Sbjct 27 ISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQATKDIFREEGFR 86
Query 44 DWFRGTFATICRDFPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATAS 103
++RG + P++ + F V LKS + + + +F GA+A A+
Sbjct 87 GFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSPYLSFVSGALAGCAAT 146
Query 104 AITHPFD 110
++PFD
Sbjct 147 LGSYPFD 153
Score = 31.2 bits (69), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 22/94 (23%), Positives = 38/94 (40%), Gaps = 16/94 (17%)
Query 29 MSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVVY------------VNLKSHMGV 76
M AF+ ++++ + G T+ P++GL F Y L S + +
Sbjct 171 MRSAFVDIIQSRGIRGLYNGLTPTLVEIVPYAGLQFGTYDMFKRWMMDWNRYKLSSKIPI 230
Query 77 EADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
D+ + + Q F CG A +A + HP D
Sbjct 231 NVDT----NLSSFQLFICGLGAGTSAKLVCHPLD 260
> YPR011c
Length=326
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 52/123 (42%), Gaps = 17/123 (13%)
Query 4 GAAVAIFNPISIVKTRME---SSLGSGGMSQA--------FLQLLRNERPLD-----WFR 47
G +V P+ ++KTR+ ++L S S+A QLL L+ +R
Sbjct 134 GCSVVATYPLDLIKTRLSIQTANLSSLNRSKAKSISKPPGIWQLLSETYRLEGGLRGLYR 193
Query 48 GTFATICRDFPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITH 107
G + T P+ L F VY L+ GV + + L GA++ A IT+
Sbjct 194 GVWPTSLGVVPYVALNFAVYEQLR-EFGVNSSDAQPSWKSNLYKLTIGAISGGVAQTITY 252
Query 108 PFD 110
PFD
Sbjct 253 PFD 255
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 46/114 (40%), Gaps = 4/114 (3%)
Query 1 VARGAAVAIFNPISIVKT--RMESSLGS--GGMSQAFLQLLRNERPLDWFRGTFATICRD 56
VA + + +P VK +++SS S G+ + Q+ E FRG R
Sbjct 31 VAGAVSRTVVSPFERVKILLQVQSSTTSYNRGIFSSIRQVYHEEGTKGLFRGNGLNCIRI 90
Query 57 FPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
FP+S + FVVY K + + + Q GA+ + T+P D
Sbjct 91 FPYSAVQFVVYEACKKKLFHVNGNNGQEQLTNTQRLFSGALCGGCSVVATYPLD 144
> 7297659
Length=399
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 34/61 (55%), Gaps = 6/61 (9%)
Query 12 PISIVKTRMES-SLGSGGMSQAFLQL----LRNERPLDWFRGTFATICRDFPFS-GLFFV 65
PI +VKT M++ +LG+ F+ RNE P +FRG +T+ R FP + FFV
Sbjct 245 PIDVVKTHMQADALGANAKYNGFIDCAMKGFRNEGPQYFFRGLNSTLIRAFPMNAACFFV 304
Query 66 V 66
V
Sbjct 305 V 305
Score = 27.3 bits (59), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 9/73 (12%)
Query 1 VARGAAVAIFNPISIVKTRME--SSLGSG----GMSQAFLQLLRNERPLDWFRGTFATIC 54
VA+G A P+ + KTR++ + + SG G +++ E F+G ATI
Sbjct 144 VAQGFVCA---PMELAKTRLQLSTQVDSGIKFTGPIHCLKYIVKTEGIRGAFKGLTATIL 200
Query 55 RDFPFSGLFFVVY 67
RD P +FV +
Sbjct 201 RDIPGFASYFVSF 213
> CE01348
Length=702
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 56/122 (45%), Gaps = 18/122 (14%)
Query 6 AVAIFNPISIVKTRMESSLGSG-----GMSQAFLQLLRNERPLDWFRGTFATICRDFPFS 60
A + P ++KTR++ + +G G+ +L++ E P+ ++GT A +CR P
Sbjct 570 AAGLVTPADVIKTRLQVAARAGQTTYNGVIDCARKLIKEEGPMSLWKGTAARVCRSSPQF 629
Query 61 GLFFVVYVNLKSHMGVE-ADSR----ERVSCYALQN--------FGCGAVAAATASAITH 107
+ + Y L+ V+ A SR E + +Q+ G +AAAT S I H
Sbjct 630 AVTLLTYEVLQRLFYVDFAGSRPTGSELATTKTIQDESSTNPDHVGGYKLAAATFSGIEH 689
Query 108 PF 109
F
Sbjct 690 KF 691
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Query 7 VAIFNPISIVKTRMESS--LGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFF 64
V NP+ IVK R++++ + G +L+ L ++G+ A RD PFS ++F
Sbjct 478 VVFTNPLEIVKIRLQTAGEVQQAGKKIGVFTVLKELGFLGLYKGSRACFLRDIPFSAIYF 537
Query 65 VVYVNLK 71
Y + K
Sbjct 538 PAYAHAK 544
> SPCC1442.03
Length=338
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/116 (25%), Positives = 48/116 (41%), Gaps = 14/116 (12%)
Query 6 AVAIFNPISIVKTRME-----------SSLGSGGMSQAFLQLLRNERPLDWFRGTFATIC 54
A ++ P ++KTR++ S+ A Q+ + E +F G ATI
Sbjct 124 ASVVYVPSEVLKTRLQLQGRYNNPHFQSNYNYPSFRGAVKQIAKQEGMKTFFYGYRATIL 183
Query 55 RDFPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
RD PFSG + Y L+ E ++ + G++A A A +T P D
Sbjct 184 RDIPFSGFQLLFYEKLRQVAQKECGQKD---IGVFRELITGSLAGAGAGFLTTPLD 236
> 7300617
Length=296
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 44/84 (52%), Gaps = 3/84 (3%)
Query 6 AVAIFNPISIVKTR-MESSLGS-GGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLF 63
A + PI +VKTR M ++ G G+ F++ + E PL +F+G ++ R P + L
Sbjct 207 ATTLTQPIDVVKTRRMNAAPGEYSGLGDVFVKTSK-EGPLAFFKGYVPSLSRLLPHTVLL 265
Query 64 FVVYVNLKSHMGVEADSRERVSCY 87
F+ L++H G + ++ + Y
Sbjct 266 FLGLEYLRTHFGYLPEPKQTGAFY 289
> At1g79900
Length=296
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 47/113 (41%), Gaps = 7/113 (6%)
Query 1 VARGAAVAIF-NPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPF 59
VA GA ++ P+ ++K R++ G +LR + +RG T+ RD P
Sbjct 114 VATGAVQSLLLTPVELIKIRLQLQQTKSGPITLAKSILRRQGLQGLYRGLTITVLRDAPA 173
Query 60 SGLFFVV--YVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
GL+F YV + H G +E + + G +A + +P D
Sbjct 174 HGLYFWTYEYVRERLHPGCRKTGQENLRTMLV----AGGLAGVASWVACYPLD 222
> CE10358
Length=294
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 35/66 (53%), Gaps = 6/66 (9%)
Query 46 FRGTFATICRDFPFSGLFFVVYVNLKSHMGVEAD-SRERVSCYALQNFGCGAVAAATASA 104
+RG AT+ R P++ + F + K + V+ + SR V Y G++AA TA+
Sbjct 72 YRGNSATMARVVPYASMQFAAFEQYKKLLKVDENGSRTPVKRYI-----TGSLAATTATM 126
Query 105 ITHPFD 110
IT+P D
Sbjct 127 ITYPLD 132
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/116 (23%), Positives = 44/116 (37%), Gaps = 14/116 (12%)
Query 1 VARGAAVAIFNPISIVKTRME--SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFP 58
+A A I P+ K R+ S L + F++ + +RG + TI P
Sbjct 119 LAATTATMITYPLDTAKARLSVSSKLQYSSLKHVFVKTYKEGGIQLLYRGIYPTILGVIP 178
Query 59 FSGLFFVVYVNLK----SHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
++G F Y LK H G +S R+ G +A + ++P D
Sbjct 179 YAGSSFFTYETLKIMYRDHRGEVENSYYRML--------FGMLAGLIGQSSSYPLD 226
> Hs5453918
Length=307
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 54/121 (44%), Gaps = 17/121 (14%)
Query 1 VARGAAVAIFNPISIVKTRMESS-----------LGSGGMSQAFLQLLRNERPLDWFRGT 49
VA V + P+ +V TR++ G+ AF Q++R+E + GT
Sbjct 110 VAGVVNVLLTTPLWVVNTRLKLQGAKFRNEDIVPTNYKGIIDAFHQIIRDEGISALWNGT 169
Query 50 FATICRDFPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPF 109
F ++ F + F+ Y LK + ++R+ +L F GAVA A A+ +T+P
Sbjct 170 FPSLLLVFN-PAIQFMFYEGLKRQL-----LKKRMKLSSLDVFIIGAVAKAIATTVTYPL 223
Query 110 D 110
Sbjct 224 Q 224
> Hs18590154
Length=94
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 38/69 (55%), Gaps = 4/69 (5%)
Query 12 PISIVKTRMESS---LGSGGMSQAFLQ-LLRNERPLDWFRGTFATICRDFPFSGLFFVVY 67
P+++V+TRM++ GS + LQ +L + L +RG T+ + P G+ +VVY
Sbjct 26 PLTLVRTRMQAQDTVEGSNPTMRGVLQRILAQQGWLGLYRGMTPTLLKVLPAGGISYVVY 85
Query 68 VNLKSHMGV 76
+K +G+
Sbjct 86 EAMKKTLGI 94
> At4g39460
Length=330
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 54/110 (49%), Gaps = 13/110 (11%)
Query 5 AAVAIFNPISIVKTRMESSLGSGGMSQAFLQLLRNERPLDWFRGTFAT----ICRDFPFS 60
AA I P +VK RM++ G + + ++++ ++ + FRG +A + RD PF
Sbjct 148 AASLIRVPTEVVKQRMQT--GQFTSAPSAVRMIASK---EGFRGLYAGYRSFLLRDLPFD 202
Query 61 GLFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+ F +Y L +G + +R +S +N GA A A A+T P D
Sbjct 203 AIQFCIYEQL--CLGYKKAARRELS--DPENALIGAFAGALTGAVTTPLD 248
> Hs13899342
Length=315
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Query 35 QLLRNERPLDWFRGTFATICRDFPFSGLFFVVYVNLKSHMGVEADSRERVSCYALQNFGC 94
+LLR + +RG AT+ RD PFS ++F ++ NL + E + + +F
Sbjct 172 ELLRTQGLAGLYRGLGATLLRDIPFSIIYFPLFANLNNLGFNELAGKASFA----HSFVS 227
Query 95 GAVAAATASAITHPFD 110
G VA + A+ P D
Sbjct 228 GCVAGSIAAVAVTPLD 243
> Hs4507009
Length=325
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 49/105 (46%), Gaps = 6/105 (5%)
Query 8 AIFNPISIVKTRMES--SLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFV 65
I NP ++K RM++ SL G M +F+ + + E +RG T R G+
Sbjct 157 TIANPTDVLKIRMQAQGSLFQGSMIGSFIDIYQQEGTRGLWRGVVPTAQRAAIVVGVELP 216
Query 66 VYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
VY K H+ + + + + + +F CG +A A AS +P D
Sbjct 217 VYDITKKHLILSGMMGDTILTHFVSSFTCG-LAGALAS---NPVD 257
Score = 31.6 bits (70), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 45/109 (41%), Gaps = 15/109 (13%)
Query 12 PISIVKTRME----------SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSG 61
P+ + KTR++ + GM A ++ + E L + G + R +
Sbjct 60 PVDLTKTRLQVQGQSIDARFKEIKYRGMFHALFRICKEEGVLALYSGIAPALLRQASYGT 119
Query 62 LFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+ +Y +LK + VE E + L N CG V+ +S I +P D
Sbjct 120 IKIGIYQSLK-RLFVERLEDETL----LINMICGVVSGVISSTIANPTD 163
Score = 30.8 bits (68), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 32/68 (47%), Gaps = 7/68 (10%)
Query 11 NPISIVKTRMESSLG-------SGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLF 63
NP+ +V+TRM + G L++ ++E ++G + R P++ +F
Sbjct 254 NPVDVVRTRMMNQRAIVGHVDLYKGTVDGILKMWKHEGFFALYKGFWPNWLRLGPWNIIF 313
Query 64 FVVYVNLK 71
F+ Y LK
Sbjct 314 FITYEQLK 321
> Hs13259543
Length=322
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 49/105 (46%), Gaps = 6/105 (5%)
Query 8 AIFNPISIVKTRMES--SLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFV 65
I NP ++K RM++ SL G M +F+ + + E +RG T R G+
Sbjct 154 TIANPTDVLKIRMQAQGSLFQGSMIGSFIDIYQQEGTRGLWRGVVPTAQRAAIVVGVELP 213
Query 66 VYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
VY K H+ + + + + + +F CG +A A AS +P D
Sbjct 214 VYDITKKHLILSGMMGDTILTHFVSSFTCG-LAGALAS---NPVD 254
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 45/109 (41%), Gaps = 15/109 (13%)
Query 12 PISIVKTRME----------SSLGSGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSG 61
P+ + KTR++ + GM A ++ + E L + G + R +
Sbjct 57 PVDLTKTRLQVQGQSIDARFKEIKYRGMFHALFRICKEEGVLALYSGIAPALLRQASYGT 116
Query 62 LFFVVYVNLKSHMGVEADSRERVSCYALQNFGCGAVAAATASAITHPFD 110
+ +Y +LK + VE E + L N CG V+ +S I +P D
Sbjct 117 IKIGIYQSLK-RLFVERLEDETL----LINMICGVVSGVISSTIANPTD 160
Score = 30.8 bits (68), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 32/68 (47%), Gaps = 7/68 (10%)
Query 11 NPISIVKTRMESSLG-------SGGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLF 63
NP+ +V+TRM + G L++ ++E ++G + R P++ +F
Sbjct 251 NPVDVVRTRMMNQRAIVGHVDLYKGTVDGILKMWKHEGFFALYKGFWPNWLRLGPWNIIF 310
Query 64 FVVYVNLK 71
F+ Y LK
Sbjct 311 FITYEQLK 318
> Hs13259541
Length=309
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 38/76 (50%), Gaps = 2/76 (2%)
Query 9 IFNPISIVKTR-MESSLGS-GGMSQAFLQLLRNERPLDWFRGTFATICRDFPFSGLFFVV 66
I +P+ +VKTR M S+LG L +L+ E P +++G + R ++ + FV
Sbjct 231 IASPVDVVKTRYMNSALGQYSSAGHCALTMLQKEGPRAFYKGFMPSFLRLGSWNVVMFVT 290
Query 67 YVNLKSHMGVEADSRE 82
Y LK + SRE
Sbjct 291 YEQLKRALMAACTSRE 306
Lambda K H
0.326 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40