bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2248_orf1
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
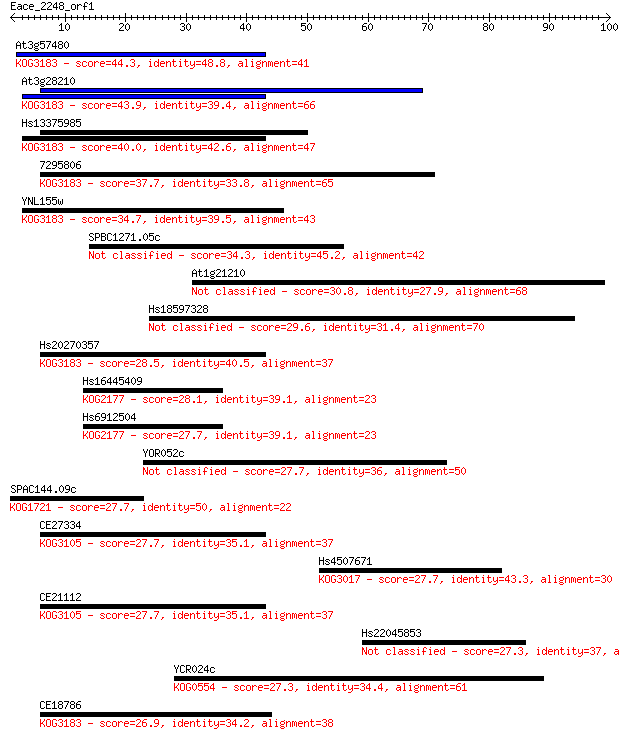
At3g57480 44.3 6e-05
At3g28210 43.9 7e-05
Hs13375985 40.0 0.001
7295806 37.7 0.006
YNL155w 34.7 0.046
SPBC1271.05c 34.3 0.062
At1g21210 30.8 0.70
Hs18597328 29.6 1.3
Hs20270357 28.5 3.5
Hs16445409 28.1 4.6
Hs6912504 27.7 4.8
YOR052c 27.7 5.0
SPAC144.09c 27.7 5.1
CE27334 27.7 5.1
Hs4507671 27.7 5.6
CE21112 27.7 6.0
Hs22045853 27.3 7.6
YCR024c 27.3 7.6
CE18786 26.9 9.1
> At3g57480
Length=249
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 20/41 (48%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 2 KGRHCPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNC 42
K + CPV CRE LT + IKC C ++ C+KHR DH+C
Sbjct 97 KKKKCPVPRCRELLTFSNTIKCRDCSIDHCLKHRFGPDHSC 137
> At3g28210
Length=186
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 38/66 (57%), Gaps = 3/66 (4%)
Query 6 CPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNC--ELLQSSKSERKRNSLFRQ-LS 62
CPVK C+E LT + + C +C V+ C+KHR DH C +++ ++ + + N F + LS
Sbjct 103 CPVKRCKEILTFANNLTCKYCGVKFCLKHRFPTDHVCNKKIINTAGTSSRWNERFMEALS 162
Query 63 SSASKG 68
KG
Sbjct 163 LRNQKG 168
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Query 3 GRHCPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNC 42
G HC C+ L C C++ C++HR + HNC
Sbjct 13 GEHCQDPDCK--LLDFLPFTCDGCKLVFCLEHRSYKSHNC 50
> Hs13375985
Length=268
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 6 CPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNCELLQSSK 49
C K C ER + A+ CP+C+ C++HR + DH CE L+ K
Sbjct 64 CSFKDCAER--ELVAVICPYCEKNFCLRHRHQSDHECEKLEIPK 105
Score = 32.0 bits (71), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query 3 GRHCPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNC 42
G+HC V+ CR+R C C C++HR +E H C
Sbjct 7 GQHCQVEHCRQR--DFLPFVCDDCSGIFCLEHRSRESHGC 44
> 7295806
Length=253
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 37/69 (53%), Gaps = 7/69 (10%)
Query 6 CPVKGC-RERLTPISAIKCPHCQVEVCIKHRLKEDHNCELLQSSKSERKR---NSLFRQL 61
C KGC R+ L P++ C C++ C++HR DH+C+ + +S + R S+F+
Sbjct 96 CNAKGCKRKELIPVT---CSQCRLNFCLRHRHTSDHDCKPVPASSTTSSRGGFQSIFKTS 152
Query 62 SSSASKGVK 70
S S S +
Sbjct 153 SDSRSMAAQ 161
> YNL155w
Length=274
Score = 34.7 bits (78), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 3 GRHCPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNCELL 45
G+HC CR+ C C + C HRLKEDH+C L
Sbjct 15 GKHCAY--CRQ--LDFLPFHCSFCNEDFCSNHRLKEDHHCRWL 53
> SPBC1271.05c
Length=215
Score = 34.3 bits (77), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 22/43 (51%), Gaps = 1/43 (2%)
Query 14 RLTPISAIKCPHCQVEVCIKHRLKEDHNC-ELLQSSKSERKRN 55
R+T A C HC C HRL EDH+C L K E +RN
Sbjct 157 RITLRLAGNCLHCNGRFCAAHRLMEDHDCVALFSLRKEEHERN 199
> At1g21210
Length=738
Score = 30.8 bits (68), Expect = 0.70, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 33/70 (47%), Gaps = 2/70 (2%)
Query 31 CIKHRLKEDHNCELLQSSKSERKRNSLFRQLSSSASKGVKMKWHSESTTNSA--GVQKDR 88
CI+H++K + EL Q + L ++LS + V +K +E A G ++R
Sbjct 355 CIEHKMKNTKDTELRQQFFEQNGGGMLMQRLSGAGPSNVDVKIFTEEGMKEATDGYDENR 414
Query 89 KEKKGGMSTI 98
+GG T+
Sbjct 415 ILGQGGQGTV 424
> Hs18597328
Length=647
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 37/80 (46%), Gaps = 11/80 (13%)
Query 24 PHCQVEVCIKHRLKEDHNC----------ELLQSSKSERKRNSLFRQLSSSASKGVKMKW 73
PH +E K +LKE+ N EL + + + R LF Q++ +K +W
Sbjct 506 PHKSLEEVFKAKLKENRNNDRKRAKEYKKELEEMKQRIQTRPYLFEQVAKDLAKKEAEQW 565
Query 74 HSESTTNSAGVQKDRKEKKG 93
+ + T AG+++D KG
Sbjct 566 YLD-TLKQAGLEEDFVRNKG 584
> Hs20270357
Length=257
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 19/37 (51%), Gaps = 2/37 (5%)
Query 6 CPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNC 42
C GCR+R + + C C CIKHR DH+C
Sbjct 100 CERAGCRQRE--MMKLTCERCSRNFCIKHRHPLDHDC 134
> Hs16445409
Length=685
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 9/23 (39%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 13 ERLTPISAIKCPHCQVEVCIKHR 35
E + PI+A +CP C+ + + HR
Sbjct 46 ESIEPITAFQCPTCRYVISLNHR 68
> Hs6912504
Length=715
Score = 27.7 bits (60), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 9/23 (39%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 13 ERLTPISAIKCPHCQVEVCIKHR 35
E + PI+A +CP C+ + + HR
Sbjct 46 ESIEPITAFQCPTCRYVISLNHR 68
> YOR052c
Length=150
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 3/51 (5%)
Query 23 CPHCQVEVCIKHRLKEDHNCELLQSSKSE-RKRNSLFRQLSSSASKGVKMK 72
C C+ C KHRL E+H C L S K + +RN+ +L + +K K++
Sbjct 101 CNFCKGHFCSKHRLMENHACNGLTSCKEQLHQRNA--DKLEAEQTKAPKIQ 149
> SPAC144.09c
Length=374
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 1 RKGRHCPVKGCRERLTPISAIK 22
RK HCP++GC++ T SA+K
Sbjct 233 RKTYHCPMEGCKKSFTRSSALK 254
> CE27334
Length=518
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 19/38 (50%), Gaps = 5/38 (13%)
Query 6 CPVKGCRERLTPISAIKCPHCQVEVCIKHRLKED-HNC 42
C GC E S I+C C ++C +H + +D H C
Sbjct 482 CGFGGCSE----FSFIRCARCNCQICFEHFVVDDQHFC 515
> Hs4507671
Length=243
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 52 RKRNSLFRQLSSSASKGVKMKWHSESTTNS 81
K N L + +S AS +KM+W E TTN+
Sbjct 42 NKHNELRKAVSPPASNMLKMEWSREVTTNA 71
> CE21112
Length=527
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 19/38 (50%), Gaps = 5/38 (13%)
Query 6 CPVKGCRERLTPISAIKCPHCQVEVCIKHRLKED-HNC 42
C GC E S I+C C ++C +H + +D H C
Sbjct 491 CGFGGCSE----FSFIRCARCNCQICFEHFVVDDQHFC 524
> Hs22045853
Length=575
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 10/27 (37%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 59 RQLSSSASKGVKMKWHSESTTNSAGVQ 85
R+L+ + ++ V +WH E+ T S G Q
Sbjct 385 RRLTEAPTRSVGAQWHPEAPTRSVGAQ 411
> YCR024c
Length=492
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 33/72 (45%), Gaps = 11/72 (15%)
Query 28 VEVCIKHRLKE--DHNCELL---------QSSKSERKRNSLFRQLSSSASKGVKMKWHSE 76
VE IKH +K D+ ELL ++ SE N +Q+ + + KWH+
Sbjct 272 VETTIKHIIKACIDNQQELLPKQFISSQENNASSELSINQETQQIKTRWEDLINEKWHNI 331
Query 77 STTNSAGVQKDR 88
+ TN+ + K R
Sbjct 332 TYTNAIEILKKR 343
> CE18786
Length=202
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Query 6 CPVKGCRERLTPISAIKCPHCQVEVCIKHRLKEDHNCE 43
C V C+++ + A+ C C+ C+ HR + DH+CE
Sbjct 98 CTVPKCKKKE--LVAMNCSKCRNNYCLSHRHERDHSCE 133
Lambda K H
0.315 0.127 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40