bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2287_orf1
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
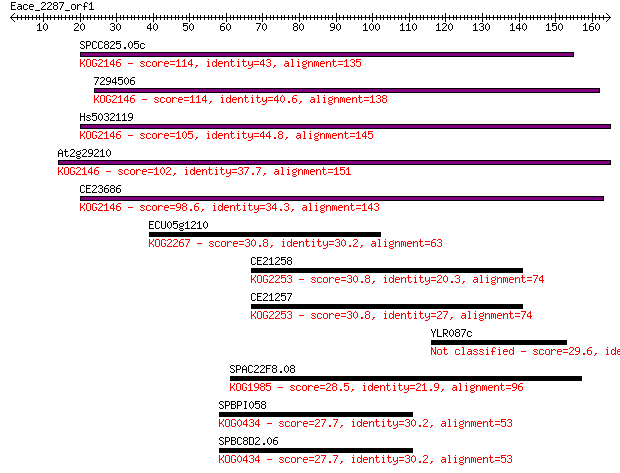
SPCC825.05c 114 7e-26
7294506 114 7e-26
Hs5032119 105 5e-23
At2g29210 102 3e-22
CE23686 98.6 5e-21
ECU05g1210 30.8 1.1
CE21258 30.8 1.2
CE21257 30.8 1.4
YLR087c 29.6 2.7
SPAC22F8.08 28.5 6.0
SPBPI058 27.7 9.8
SPBC8D2.06 27.7 9.8
> SPCC825.05c
Length=301
Score = 114 bits (286), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 58/138 (42%), Positives = 88/138 (63%), Gaps = 6/138 (4%)
Query 20 MSGNFYKGTSKDQTPFFKDKDSQLIAQRKWPAIFEQEVDMSKVNVEVMKAWINHKITELL 79
MSG FYKG + +Q F D +L+ K+PA ++ +VDM KVN+EV+K WI ++ EL+
Sbjct 1 MSG-FYKGVAAEQETLFTTADKKLMRSTKFPASYDTKVDMKKVNIEVLKPWIATRLNELI 59
Query 80 GFEDDIVISYCLSQLVPEEALAADV---DERKNYLCPKKLAISLTGFVGKQATHFVRELW 136
GFED++VI++ L EEA+ A + ++ L P+K+ ++LTGF+ AT F ELW
Sbjct 60 GFEDEVVINFVYGML--EEAVEASKTSDSQNESTLDPRKVQLNLTGFLESNATAFTEELW 117
Query 137 KLLLSAQNTPTGIPPEFL 154
L++SA GIP +F+
Sbjct 118 SLIISASQNQYGIPEKFI 135
> 7294506
Length=954
Score = 114 bits (286), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 56/139 (40%), Positives = 89/139 (64%), Gaps = 12/139 (8%)
Query 24 FYKGTSKDQTPFFKDKDSQLIAQRKWPAIFEQEVDMSKVNVEVMKAWINHKITELLGFED 83
+ GT++ Q F DK+ +L+ Q K+ + VDMSKV ++V++ WI+ KIT++L ED
Sbjct 2 MFTGTNQQQDTRFSDKEKKLMKQMKFGDCLNKRVDMSKVKLDVLRPWISKKITDILHIED 61
Query 84 DIVISYCLSQLVPEEALAADVDERKNYLCPKKLAISLTGFV-GKQATHFVRELWKLLLSA 142
D+V+ + +QL E + Y CPKK+ I++TGF+ G+ A F+ ELW LLLSA
Sbjct 62 DVVVEFVYNQL-----------EEEKYPCPKKMQINMTGFLNGRNARQFMGELWALLLSA 110
Query 143 QNTPTGIPPEFLENRRLEM 161
Q + +GIP EF++ ++ E+
Sbjct 111 QESDSGIPAEFIQQKKDEI 129
> Hs5032119
Length=820
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 65/146 (44%), Positives = 92/146 (63%), Gaps = 13/146 (8%)
Query 20 MSGNFYKGTSKDQTPFFKDKDSQLIAQRKWPAIFEQEVDMSKVNVEVMKAWINHKITELL 79
M F++GTS +Q F +K +L+ Q K+ E++VDMSKVN+EV+K WI ++TE+L
Sbjct 1 MDAGFFRGTSAEQDNRFSNKQKKLLKQLKFAECLEKKVDMSKVNLEVIKPWITKRVTEIL 60
Query 80 GFEDDIVISYCLSQLVPEEALAADVDERKNYLCPKKLAISLTGFV-GKQATHFVRELWKL 138
GFEDD+VI + +QL E KN K + I+LTGF+ GK A F+ ELW L
Sbjct 61 GFEDDVVIEFIFNQL-----------EVKNPDS-KMMQINLTGFLNGKNAREFMGELWPL 108
Query 139 LLSAQNTPTGIPPEFLENRRLEMERK 164
LLSAQ GIP FLE ++ E++++
Sbjct 109 LLSAQENIAGIPSAFLELKKEEIKQR 134
> At2g29210
Length=891
Score = 102 bits (255), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 57/154 (37%), Positives = 88/154 (57%), Gaps = 15/154 (9%)
Query 14 VCSEIEMSG---NFYKGTSKDQTPFFKDKDSQLIAQRKWPAIFEQEVDMSKVNVEVMKAW 70
C E E N GTS +Q F +K ++L+ +K+ E VD++KV ++VMK W
Sbjct 12 ACQEDETEHVPINGNNGTSAEQDTRFSNKQAKLMKSQKFAPELENLVDITKVKMDVMKPW 71
Query 71 INHKITELLGFEDDIVISYCLSQLVPEEALAADVDERKNYLCPKKLAISLTGFVGKQATH 130
I ++TELLG ED+++I++ L + K++ I+LTGF+ K
Sbjct 72 IATRVTELLGIEDEVLINFIYGLLD------------GKVVNGKEIQITLTGFMEKNTGK 119
Query 131 FVRELWKLLLSAQNTPTGIPPEFLENRRLEMERK 164
F++ELW LLLSAQN P+G+P +FL+ R E ++K
Sbjct 120 FMKELWTLLLSAQNNPSGVPQQFLDARAAETKKK 153
> CE23686
Length=335
Score = 98.6 bits (244), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 85/144 (59%), Gaps = 13/144 (9%)
Query 20 MSGNFYKGTSKDQTPFFKDKDSQLIAQRKWPAIFEQEVDMSKVNVEVMKAWINHKITELL 79
M +++G + +Q F DK+ +L+ K+ EQ++D+++ N++V+K WI ++ ++L
Sbjct 1 MDAGYFRGANSEQDGRFSDKEKKLLKTMKFEPQLEQKIDLNRCNMDVIKPWITARVNDIL 60
Query 80 GFEDDIVISYCLSQLVPEEALAADVDERKNYLCPKKLAISLTGFV-GKQATHFVRELWKL 138
G EDD+V+ Y LSQ+ + KN L PK L I++TGF+ ++A FV +LW L
Sbjct 61 GMEDDVVVEYILSQI-----------DDKN-LNPKLLQINVTGFLNARRAREFVGDLWNL 108
Query 139 LLSAQNTPTGIPPEFLENRRLEME 162
L+ A + GIP + + EM+
Sbjct 109 LIEANASEDGIPASLVNQKMAEMK 132
> ECU05g1210
Length=399
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 1/64 (1%)
Query 39 KDSQLIAQRKWPAI-FEQEVDMSKVNVEVMKAWINHKITELLGFEDDIVISYCLSQLVPE 97
K ++ + WP I F + S V+ E + +I L E D +IS CLS + E
Sbjct 2 KRAKTAKEGGWPGITFYTALSKSSVDYETFRKCAERRIEMLRCEEGDGIISKCLSTSIEE 61
Query 98 EALA 101
+ L+
Sbjct 62 DILS 65
> CE21258
Length=554
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/74 (20%), Positives = 33/74 (44%), Gaps = 13/74 (17%)
Query 67 MKAWINHKITELLGFEDDIVISYCLSQLVPEEALAADVDERKNYLCPKKLAISLTGFVGK 126
++ W+ K+T+ LG ED + Q+ + P+++ + + +
Sbjct 483 IRPWVAKKVTQFLGEEDKSFCDFICDQI-------------EKQATPQEILKDVAVIIDE 529
Query 127 QATHFVRELWKLLL 140
A FV ++W+LL+
Sbjct 530 DAEQFVIKMWRLLI 543
> CE21257
Length=712
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 35/78 (44%), Gaps = 21/78 (26%)
Query 67 MKAWINHKITELLGFED----DIVISYCLSQLVPEEALAADVDERKNYLCPKKLAISLTG 122
++ W+ K+T+ LG ED D + Q P+E L K +A+
Sbjct 641 IRPWVAKKVTQFLGEEDKSFCDFICDQIEKQATPQEIL-------------KDVAV---- 683
Query 123 FVGKQATHFVRELWKLLL 140
+ + A FV ++W+LL+
Sbjct 684 IIDEDAEQFVIKMWRLLI 701
> YLR087c
Length=2958
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 116 LAISLTGF---VGKQATHFVRELWKLLLSAQNTPTGIPPE 152
L S+T F V +T+FV EL LLLS NT G+ PE
Sbjct 1924 LDTSITYFGVLVPVASTYFVFELHMLLLSLTNTNNGMLPE 1963
> SPAC22F8.08
Length=926
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 21/96 (21%), Positives = 43/96 (44%), Gaps = 14/96 (14%)
Query 61 KVNVEVMKAWINHKITELLGFEDDIVISYCLSQLVPEEALAADVDERKNYLCPKKLAISL 120
++ V + + +T+L D + I+ L+ E+AL++ ++E ++ + K + I
Sbjct 643 RIRVLTISLPTTNSMTDLYASADQVAIAQYLTVRASEKALSSTLNEARDSIISKLVEI-- 700
Query 121 TGFVGKQATHFVRELWKLLLSAQNTPTGIPPEFLEN 156
E++K L+ QNT IP + N
Sbjct 701 ------------LEVYKKNLAGQNTGAAIPLQISTN 724
> SPBPI058
Length=1064
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query 58 DMSKVNVEVMKAWINHKITELLGFEDDIVISYCLSQLVPEEALAADVDERKNY 110
D + ++ VM WI + L+GF D+ + Y L +VP+ L ++E N+
Sbjct 679 DAATLSSNVMDRWILARCQSLIGFVDEEMKQYRLYTVVPQ--LLGLIEEMTNW 729
> SPBC8D2.06
Length=1064
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query 58 DMSKVNVEVMKAWINHKITELLGFEDDIVISYCLSQLVPEEALAADVDERKNY 110
D + ++ VM WI + L+GF D+ + Y L +VP+ L ++E N+
Sbjct 679 DAATLSSNVMDRWILARCQSLIGFVDEEMKQYRLYTVVPQ--LLGLIEEMTNW 729
Lambda K H
0.318 0.133 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2317343104
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40