bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2356_orf1
Length=80
Score E
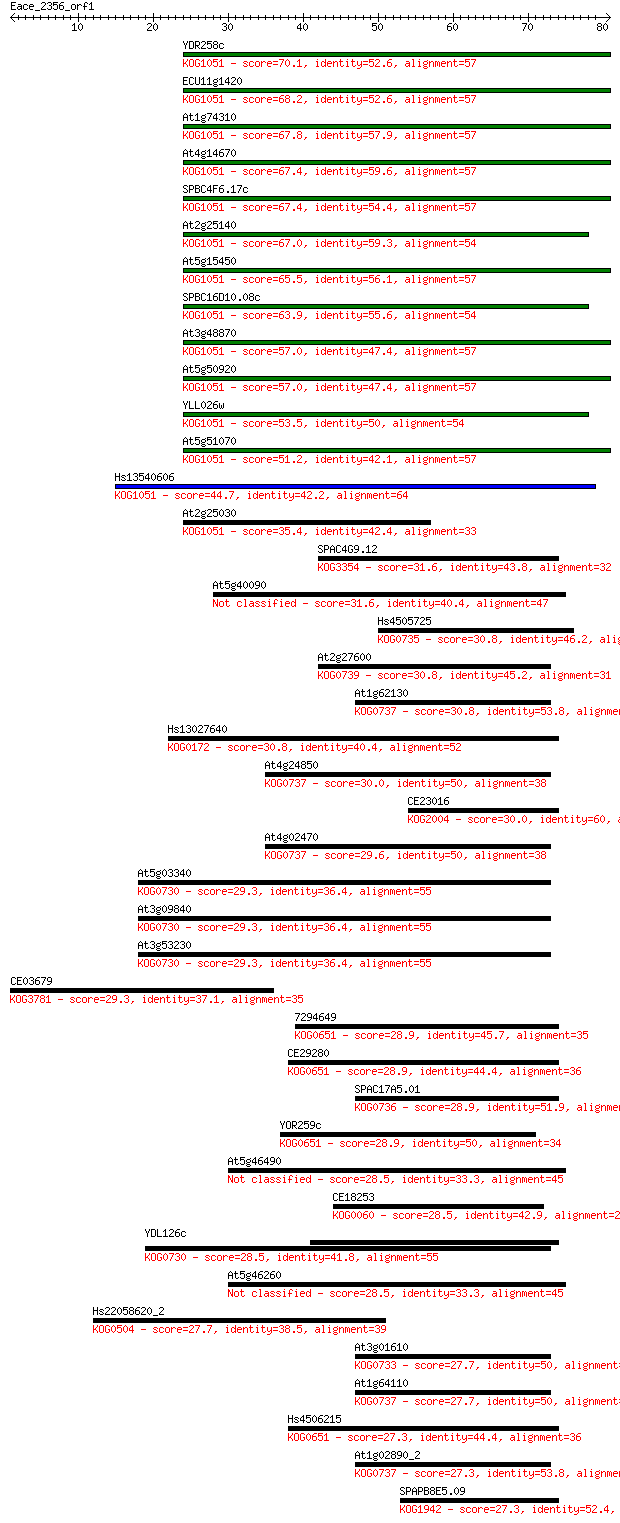
Sequences producing significant alignments: (Bits) Value
YDR258c 70.1 1e-12
ECU11g1420 68.2 3e-12
At1g74310 67.8 5e-12
At4g14670 67.4 6e-12
SPBC4F6.17c 67.4 6e-12
At2g25140 67.0 8e-12
At5g15450 65.5 3e-11
SPBC16D10.08c 63.9 7e-11
At3g48870 57.0 7e-09
At5g50920 57.0 9e-09
YLL026w 53.5 1e-07
At5g51070 51.2 4e-07
Hs13540606 44.7 5e-05
At2g25030 35.4 0.022
SPAC4G9.12 31.6 0.36
At5g40090 31.6 0.38
Hs4505725 30.8 0.57
At2g27600 30.8 0.66
At1g62130 30.8 0.67
Hs13027640 30.8 0.69
At4g24850 30.0 1.1
CE23016 30.0 1.2
At4g02470 29.6 1.3
At5g03340 29.3 1.8
At3g09840 29.3 1.8
At3g53230 29.3 1.9
CE03679 29.3 2.0
7294649 28.9 2.2
CE29280 28.9 2.4
SPAC17A5.01 28.9 2.5
YOR259c 28.9 2.6
At5g46490 28.5 2.8
CE18253 28.5 3.0
YDL126c 28.5 3.2
At5g46260 28.5 3.2
Hs22058620_2 27.7 4.8
At3g01610 27.7 5.5
At1g64110 27.7 5.6
Hs4506215 27.3 6.1
At1g02890_2 27.3 6.7
SPAPB8E5.09 27.3 7.7
> YDR258c
Length=811
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 30/57 (52%), Positives = 45/57 (78%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +A+ A++DA+ +QRAGL+ + +P+ +FMFLG +G GKTEL KA+AE +FD E N+
Sbjct 509 QDEAIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGTGKTELTKALAEFLFDDESNV 565
> ECU11g1420
Length=851
Score = 68.2 bits (165), Expect = 3e-12, Method: Composition-based stats.
Identities = 30/57 (52%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q AV A+ D++ R GL ++P+G+F+ LG +GVGKTELAKAVA E+FD+EK++
Sbjct 557 QDHAVDAIVDSILQSRVGLDDDDRPVGSFLLLGPTGVGKTELAKAVAMELFDNEKDM 613
> At1g74310
Length=911
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 33/57 (57%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q QAV AV++A+ RAGL +P G+F+FLG +GVGKTELAKA+AE++FD E L
Sbjct 574 QNQAVNAVSEAILRSRAGLGRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLL 630
> At4g14670
Length=623
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 34/57 (59%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AVKAVA A+ R GL +P G+F+FLG +GVGKTELAKA+AE++FDSE L
Sbjct 539 QDEAVKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLL 595
> SPBC4F6.17c
Length=803
Score = 67.4 bits (163), Expect = 6e-12, Method: Composition-based stats.
Identities = 31/57 (54%), Positives = 45/57 (78%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +A+KA+ADA+ + RAGL N+PL +F+FLG +GVGKT L KA+AE +FD++K +
Sbjct 507 QDEALKAIADAVRLSRAGLQNTNRPLASFLFLGPTGVGKTALTKALAEFLFDTDKAM 563
> At2g25140
Length=874
Score = 67.0 bits (162), Expect = 8e-12, Method: Composition-based stats.
Identities = 32/54 (59%), Positives = 43/54 (79%), Gaps = 0/54 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 77
Q AVK+VADA+ RAGLS N+P+ +FMF+G +GVGKTELAKA+A +F++E
Sbjct 568 QDMAVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAGYLFNTE 621
> At5g15450
Length=968
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 32/57 (56%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q AV AVA+A+ RAGLS +P+ +FMF+G +GVGKTELAKA+A MF++E+ L
Sbjct 653 QNPAVTAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVGKTELAKALASYMFNTEEAL 709
> SPBC16D10.08c
Length=905
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 30/54 (55%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 77
Q +AV AVA+A+ + RAGLS N+P+ +F+F G SG GKT L KA+A MFD E
Sbjct 589 QNEAVTAVANAIRLSRAGLSDPNQPIASFLFCGPSGTGKTLLTKALASFMFDDE 642
> At3g48870
Length=952
Score = 57.0 bits (136), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AVKA++ A+ R GL N+P+ +F+F G +GVGK+ELAKA+A F SE+ +
Sbjct 634 QDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 690
> At5g50920
Length=929
Score = 57.0 bits (136), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AVKA++ A+ R GL N+P+ +F+F G +GVGK+ELAKA+A F SE+ +
Sbjct 613 QDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAM 669
> YLL026w
Length=908
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 39/54 (72%), Gaps = 1/54 (1%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSE 77
Q A+KAV++A+ + R+GL+ +P +F+FLG SG GKTELAK VA +F+ E
Sbjct 583 QMDAIKAVSNAVRLSRSGLANPRQP-ASFLFLGLSGSGKTELAKKVAGFLFNDE 635
> At5g51070
Length=945
Score = 51.2 bits (121), Expect = 4e-07, Method: Composition-based stats.
Identities = 24/57 (42%), Positives = 37/57 (64%), Gaps = 0/57 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMFDSEKNL 80
Q +AV A++ A+ R GL ++P+ +F G +GVGKTEL KA+A F SE+++
Sbjct 632 QDEAVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGSEESM 688
> Hs13540606
Length=707
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 15 PQQQQLPRQ---QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAE 71
P +Q+L Q+ A+ V A+ + G + PL F+FLGSSG+GKTELAK A+
Sbjct 338 PLEQRLKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAK 396
Query 72 EMFDSEK 78
M K
Sbjct 397 YMHKDAK 403
> At2g25030
Length=265
Score = 35.4 bits (80), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 24 QQQAVKAVADALAIQRAGLSPKNKPLGTFMFLG 56
Q V++VADA+ +AG+S N+ + +FMF+G
Sbjct 13 QDLDVESVADAIRCSKAGISDPNRLIASFMFMG 45
> SPAC4G9.12
Length=193
Score = 31.6 bits (70), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 24/33 (72%), Gaps = 1/33 (3%)
Query 42 LSPKNKPLG-TFMFLGSSGVGKTELAKAVAEEM 73
++P N+P F+ +G +G GKT +AKAV+E++
Sbjct 6 INPTNQPYKYVFVVIGPAGSGKTTMAKAVSEKL 38
> At5g40090
Length=459
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 6/47 (12%)
Query 28 VKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF 74
+K V D LA++ NK + T GS+GVGKT LA+ + E+F
Sbjct 188 MKVVYDLLALE------VNKEVRTIGIWGSAGVGKTTLARYIYAEIF 228
> Hs4505725
Length=1283
Score = 30.8 bits (68), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 50 GTFMFLGSSGVGKTELAKAVAEEMFD 75
G + G G GK+ LAKA+ +E FD
Sbjct 593 GALLLTGGKGSGKSTLAKAICKEAFD 618
> At2g27600
Length=435
Score = 30.8 bits (68), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 42 LSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ K +P F+ G G GK+ LAKAVA E
Sbjct 158 FTGKRRPWRAFLLYGPPGTGKSYLAKAVATE 188
> At1g62130
Length=372
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVAEE 72
+P + G SG GKT LAKAVA E
Sbjct 135 QPCNGILLFGPSGTGKTMLAKAVATE 160
> Hs13027640
Length=926
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query 22 RQQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 73
R QAV+AV DA GL PK+ TF+F G+ V K A+A+ E+
Sbjct 189 RNSSQAVQAVRDAGYEISLGLMPKSIGPLTFVFTGTGNVSKG--AQAIFNEL 238
> At4g24850
Length=442
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 3/41 (7%)
Query 35 LAIQRAGLSPK---NKPLGTFMFLGSSGVGKTELAKAVAEE 72
L +QR L K KP + G G GKT LAKAVA+E
Sbjct 158 LPLQRPELFCKGELTKPCKGILLFGPPGTGKTMLAKAVAKE 198
> CE23016
Length=773
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 54 FLGSSGVGKTELAKAVAEEM 73
F G G+GKT +AKA+AE M
Sbjct 334 FTGPPGIGKTSIAKAIAESM 353
> At4g02470
Length=371
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 21/41 (51%), Gaps = 3/41 (7%)
Query 35 LAIQRAGLSPK---NKPLGTFMFLGSSGVGKTELAKAVAEE 72
L +QR L K KP + G G GKT LAKAVA E
Sbjct 87 LPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAKAVATE 127
> At5g03340
Length=843
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 18 QQLPRQQQQAVK-AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ + R+ Q+ V+ V ++ G+SP L F G G GKT LAKA+A E
Sbjct 519 ENVKRELQETVQYPVEHPEKFEKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 570
> At3g09840
Length=809
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 18 QQLPRQQQQAVK-AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ + R+ Q+ V+ V ++ G+SP L F G G GKT LAKA+A E
Sbjct 486 ENVKRELQETVQYPVEHPEKFEKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 537
> At3g53230
Length=815
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 18 QQLPRQQQQAVK-AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+ + R+ Q+ V+ V ++ G+SP L F G G GKT LAKA+A E
Sbjct 487 ENVKRELQETVQYPVEHPEKFEKFGMSPSKGVL----FYGPPGCGKTLLAKAIANE 538
> CE03679
Length=584
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 1 THHHLQMETPQPPPPQQQQLPRQQQQAVKAVADAL 35
T+H +Q+E QP P Q P ++ V+A+A +L
Sbjct 337 TNHRVQLEMDQPTPQQMATNPSKRNLLVRALARSL 371
> 7294649
Length=398
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 22/35 (62%), Gaps = 4/35 (11%)
Query 39 RAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 73
R G+SP P G ++ G G GKT LA+A+A +M
Sbjct 170 RVGISP---PKGCLLY-GPPGTGKTLLARAIASQM 200
> CE29280
Length=406
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 4/36 (11%)
Query 38 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 73
+R G++P P G +F G G GKT LA+AVA ++
Sbjct 177 KRVGITP---PKGCLLF-GPPGTGKTLLARAVASQL 208
> SPAC17A5.01
Length=948
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVAEEM 73
KP + G G GKT LAKAVA E+
Sbjct 686 KPRSGVLLYGPPGTGKTLLAKAVATEL 712
> YOR259c
Length=437
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 4/34 (11%)
Query 37 IQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVA 70
QR G+ P P G ++ G G GKT LAKAVA
Sbjct 207 FQRVGIKP---PKGVLLY-GPPGTGKTLLAKAVA 236
> At5g46490
Length=858
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 30 AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF 74
+ D +A A L ++K + GSSG+GKT +A+A+ +F
Sbjct 184 GLEDHIANMSALLDLESKEVKMVGIWGSSGIGKTTIARALFNNLF 228
> CE18253
Length=598
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 18/28 (64%), Gaps = 3/28 (10%)
Query 44 PKNKPLGTFMFLGSSGVGKTELAKAVAE 71
P+NK T + G SG+GKT L + +A+
Sbjct 405 PRNK---TLLITGDSGIGKTSLMRVIAD 429
> YDL126c
Length=835
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 18/33 (54%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Query 41 GLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 73
GLSP L F G G GKT LAKAVA E+
Sbjct 517 GLSPSKGVL----FYGPPGTGKTLLAKAVATEV 545
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 27/54 (50%), Gaps = 13/54 (24%)
Query 19 QLPRQQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEE 72
+LP + Q KA+ G+ P P G M+ G G GKT +A+AVA E
Sbjct 231 ELPLRHPQLFKAI---------GIKP---PRGVLMY-GPPGTGKTLMARAVANE 271
> At5g46260
Length=1205
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 30 AVADALAIQRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEMF 74
+ D +A A L ++K + GSSG+GKT +A+A+ +F
Sbjct 184 GLEDHIANMSALLDLESKEVKMVGIWGSSGIGKTTIARALFNNLF 228
> Hs22058620_2
Length=456
Score = 27.7 bits (60), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 25/39 (64%), Gaps = 3/39 (7%)
Query 12 PPPPQQQQLPRQQQQAVKAVADALAIQRAGLSPKNKPLG 50
PPPP +QL RQ Q++++ A +++ SP ++PLG
Sbjct 311 PPPPGMEQLGRQSQKSLEEDAKGGSVRA---SPCHEPLG 346
> At3g01610
Length=703
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVAEE 72
KP +F G G GKT+LA A+A E
Sbjct 148 KPPSGILFHGPPGCGKTKLANAIANE 173
> At1g64110
Length=821
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVAEE 72
KP + G G GKT LAKA+A+E
Sbjct 545 KPCRGILLFGPPGTGKTMLAKAIAKE 570
> Hs4506215
Length=389
Score = 27.3 bits (59), Expect = 6.1, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 22/36 (61%), Gaps = 4/36 (11%)
Query 38 QRAGLSPKNKPLGTFMFLGSSGVGKTELAKAVAEEM 73
QR G+ P P G ++ G G GKT LA+AVA ++
Sbjct 160 QRVGIIP---PKGCLLY-GPPGTGKTLLARAVASQL 191
> At1g02890_2
Length=563
Score = 27.3 bits (59), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 47 KPLGTFMFLGSSGVGKTELAKAVAEE 72
KP + G G GKT LAKAVA E
Sbjct 298 KPTKGILLFGPPGTGKTMLAKAVATE 323
> SPAPB8E5.09
Length=456
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 11/21 (52%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 53 MFLGSSGVGKTELAKAVAEEM 73
+F G +G GKT LA A+A+E+
Sbjct 68 LFAGGAGTGKTALALAIAQEL 88
Lambda K H
0.312 0.127 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1165602088
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40