bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2418_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
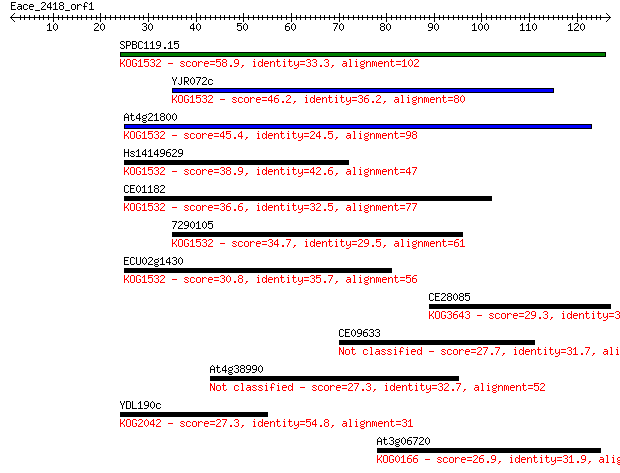
SPBC119.15 58.9 2e-09
YJR072c 46.2 1e-05
At4g21800 45.4 2e-05
Hs14149629 38.9 0.002
CE01182 36.6 0.012
7290105 34.7 0.047
ECU02g1430 30.8 0.56
CE28085 29.3 1.7
CE09633 27.7 5.3
At4g38990 27.3 7.7
YDL190c 27.3 7.8
At3g06720 26.9 8.7
> SPBC119.15
Length=367
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 62/111 (55%), Gaps = 9/111 (8%)
Query 24 DFIEALLKDER---------YLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCL 74
+F +A+ KDE Y+GSL S +L L EFY + +S S+V+ +G+D +
Sbjct 197 EFQQAVTKDEGMSSEGATSGYMGSLVNSMSLMLEEFYRHLDFVSCSSVTGEGMDDFLEAV 256
Query 75 KKLRREYEDNFIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNSSDG 125
K +EYE+ ++P + + +E ++ K ++++ QLS+ + D SK+ G
Sbjct 257 KAKVKEYEEEYVPEMERMKEIQRQTKERQKEAQLSKLMKDMHVSKDKEDVG 307
> YJR072c
Length=385
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 8/84 (9%)
Query 35 YLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDNFIPYLLQQRE 94
Y+ SL S +L L EFY ++ + VS+ + G D C+ K EY+ Y Q+RE
Sbjct 213 YMSSLVNSMSLMLEEFYSQLDVVGVSSFTGDGFDEFMQCVDKKVDEYDQ----YYKQERE 268
Query 95 KA----KKKKLQKQQQQLSRFLAD 114
KA KKK+ ++Q+ L+ + D
Sbjct 269 KALNLKKKKEEMRKQKSLNGLMKD 292
> At4g21800
Length=379
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 52/98 (53%), Gaps = 0/98 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDN 84
F A+ D Y +L S +L+L EFY I+++ VSA+S G+D + ++ EY +
Sbjct 231 FQAAIQSDNSYTATLANSLSLSLYEFYRNIRSVGVSAISGAGMDGFFKAIEASAEEYMET 290
Query 85 FIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNS 122
+ L ++ ++ + ++++ ++ + D SS+ +
Sbjct 291 YKADLDMRKADKERLEEERKKHEMEKLRKDMESSQGGT 328
> Hs14149629
Length=374
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLW 71
F +AL ++ Y+ +L RS +L L EFY ++ + VSAV GLD L+
Sbjct 209 FQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSAVLGTGLDELF 255
> CE01182
Length=355
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 45/79 (56%), Gaps = 5/79 (6%)
Query 25 FIEALLKDER--YLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYE 82
F EAL +D R Y+ L RS +L L EFY ++T+ VS+ + +G + + + + Y+
Sbjct 220 FDEAL-EDARSSYMNDLSRSLSLVLDEFYCGLKTVCVSSATGEGFEDVMTAIDESVEAYK 278
Query 83 DNFIPYLLQQREKAKKKKL 101
++P + ++ A+KK L
Sbjct 279 KEYVP--MYEKVLAEKKLL 295
> 7290105
Length=382
Score = 34.7 bits (78), Expect = 0.047, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 35 YLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRREYEDNFIPYLLQQRE 94
++ +L R+ +L L FYE ++T VSA + G +L + + EYE ++ P ++R+
Sbjct 223 FVSNLTRTMSLTLDTFYENLRTCGVSAKTGVGFTQLLTKILESVDEYETDYKPVYEKKRQ 282
Query 95 K 95
+
Sbjct 283 E 283
> ECU02g1430
Length=270
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 32/56 (57%), Gaps = 1/56 (1%)
Query 25 FIEALLKDERYLGSLGRSTALALCEFYEKIQTMSVSAVSMQGLDRLWGCLKKLRRE 80
F E+L +D+ + LG S AL EFY I+T+SVS+ + G +G + ++ E
Sbjct 211 FRESLNEDDMFSPVLG-SMALHFEEFYNSIRTVSVSSYTGYGKSDFFGAVDQMLDE 265
> CE28085
Length=506
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 89 LLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNSSDGT 126
L+ +REK +K Q+Q+ ++ F A +++ NNS + T
Sbjct 344 LVLRREKRRKAAEQQQRNEMPMFNASPKAANNNSYEMT 381
> CE09633
Length=710
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 70 LWGCLKKLRREYEDNFIPYLLQQREKAKK-KKLQKQQQQLSR 110
LW +KK + ++ P + RE+ K+ +KL K+QQ++ +
Sbjct 502 LWLAMKKFPARFAPSYDPISMWTREEVKEIQKLPKEQQEIMK 543
> At4g38990
Length=494
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 26/54 (48%), Gaps = 12/54 (22%)
Query 43 TALALCEFYEKIQTMSVSAVSMQGLDR--LWGCLKKLRREYEDNFIPYLLQQRE 94
T LA+C FY S+ G + LWG R +D++I YL++ R+
Sbjct 220 TKLAVCPFY----------CSVNGYEDELLWGAAWLRRATGKDSYIKYLVENRQ 263
> YDL190c
Length=961
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 20/40 (50%), Gaps = 9/40 (22%)
Query 24 DFIEALLKDERYLGS---------LGRSTALALCEFYEKI 54
+FI A+ KDER LGR T LA EF EK+
Sbjct 818 EFISAVAKDERSFNRNLFVRAVDILGRKTGLASPEFIEKL 857
> At3g06720
Length=532
Score = 26.9 bits (58), Expect = 8.7, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 78 RREYEDNFIPYLLQQREKAKKKKLQKQQQQLSRFLADSRSSKNNSSD 124
RR EDN + +RE++ KK ++ Q L F + S +S + D
Sbjct 26 RRRREDNMVEIRKSKREESLMKKRREGMQALQGFPSASAASVDKKLD 72
Lambda K H
0.318 0.132 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40