bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
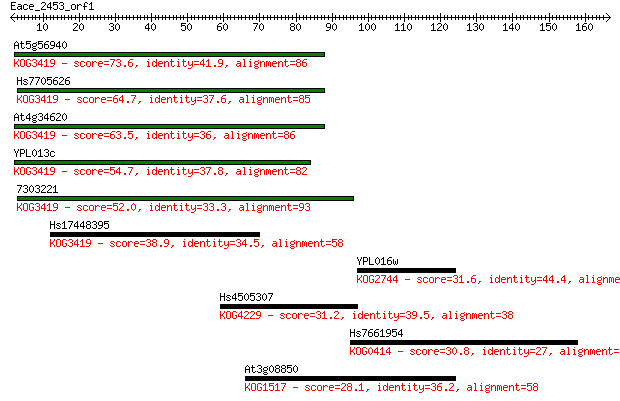
Query= Eace_2453_orf1
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
At5g56940 73.6 2e-13
Hs7705626 64.7 8e-11
At4g34620 63.5 2e-10
YPL013c 54.7 9e-08
7303221 52.0 6e-07
Hs17448395 38.9 0.004
YPL016w 31.6 0.83
Hs4505307 31.2 0.99
Hs7661954 30.8 1.4
At3g08850 28.1 7.4
> At5g56940
Length=135
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/86 (41%), Positives = 51/86 (59%), Gaps = 2/86 (2%)
Query 2 RIRMQVMGTKGRRFFKIVAANQRDPRDGKHMEVLGSYVSRAATDNKGVSELRLRFSRVKF 61
RIR+ G K R FF+++AA+ R PRDGKH+EVLG + D G + L+F R+K+
Sbjct 4 RIRLSRFGCKNRPFFRVMAADSRSPRDGKHLEVLGYFNPLPGQD--GGKRMGLKFDRIKY 61
Query 62 WLAAGASISAAAAKALGAAALIPLQP 87
WL+ GA S + L + L+P P
Sbjct 62 WLSVGAQPSDPVQRLLFRSGLLPPPP 87
> Hs7705626
Length=137
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 47/85 (55%), Gaps = 2/85 (2%)
Query 3 IRMQVMGTKGRRFFKIVAANQRDPRDGKHMEVLGSYVSRAATDNKGVSELRLRFSRVKFW 62
IR+ + G R F++IVAA+ + PRDG+ +E LGSY ++ G + L R++ W
Sbjct 19 IRLALGGCTNRPFYRIVAAHNKCPRDGRFVEQLGSY--DPLPNSHGEKLVALNLDRIRHW 76
Query 63 LAAGASISAAAAKALGAAALIPLQP 87
+ GA +S K LG A PL P
Sbjct 77 IGCGAHLSKPMEKLLGLAGFFPLHP 101
> At4g34620
Length=113
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 50/86 (58%), Gaps = 2/86 (2%)
Query 2 RIRMQVMGTKGRRFFKIVAANQRDPRDGKHMEVLGSYVSRAATDNKGVSELRLRFSRVKF 61
+IR+ +G K R F+++V A+++ RDGK +EVLG Y ++ + L+F R+K+
Sbjct 4 KIRLARLGCKHRPFYRVVVADEKSRRDGKQIEVLGFYDPLQGKED--ADRVSLKFDRIKY 61
Query 62 WLAAGASISAAAAKALGAAALIPLQP 87
WL+ GA + L A LIP +P
Sbjct 62 WLSVGAQPTDTVESMLFRAGLIPPKP 87
> YPL013c
Length=121
Score = 54.7 bits (130), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 44/90 (48%), Gaps = 8/90 (8%)
Query 2 RIRMQVMGTKGRRFFKIVAANQRDPRDGKHMEVLGSYVSRAATDNK--------GVSELR 53
RIR+ G K + IV AN R RD K +EVLG+YV + K + +++
Sbjct 7 RIRLARFGRKNSPVYNIVVANSRKARDAKPIEVLGTYVPVPSPVTKRELKRGVVPIKDVK 66
Query 54 LRFSRVKFWLAAGASISAAAAKALGAAALI 83
L F R K+W+ GA S K L A ++
Sbjct 67 LDFDRTKYWIGVGAQPSETVTKLLRKAGIL 96
> 7303221
Length=129
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 48/95 (50%), Gaps = 4/95 (4%)
Query 3 IRMQVMGTKGRRFFKIVAANQRDPRDGKHMEVLGSYVSRAATDNKGVSELRLRFSRVKFW 62
IR +G R F+ IV +R + +E +GS+ N+ + + L R+++W
Sbjct 20 IRFVRLGCTNRPFYHIVVMERRKNQHQPVIEQVGSFDPLPNDYNERL--VALNTERIRYW 77
Query 63 LAAGASISAAAAKALGAAALIPLQPPLY--GWRCR 95
L GA +S AA+ LG A L+P+ P Y WR R
Sbjct 78 LGKGAHLSTPAAELLGIAGLLPIHPRTYMTAWRNR 112
> Hs17448395
Length=215
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 32/58 (55%), Gaps = 2/58 (3%)
Query 12 GRRFFKIVAANQRDPRDGKHMEVLGSYVSRAATDNKGVSELRLRFSRVKFWLAAGASI 69
R F+ IVAA+ + PRDG +E LGSY S ++ G + + L R++ W+ +
Sbjct 159 NRPFYTIVAAHSKCPRDGLLVEQLGSYDS--FPNSHGENLIALNLDRIQHWIGCATHL 214
> YPL016w
Length=1314
Score = 31.6 bits (70), Expect = 0.83, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 97 RLQELLQEQQQQQQMLAERELRSFFKL 123
R Q+LL++QQ+QQ++L E +LR +++
Sbjct 621 RQQKLLEDQQRQQKLLLETKLRQQYEI 647
> Hs4505307
Length=2215
Score = 31.2 bits (69), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 0/38 (0%)
Query 59 VKFWLAAGASISAAAAKALGAAALIPLQPPLYGWRCRG 96
+ WL+ G S AA G A L P P W CRG
Sbjct 1525 CRVWLSLGCSDLGCAAPHSGWAGLTPAGPCSPCWSCRG 1562
> Hs7661954
Length=1401
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Query 95 RGRLQELLQEQQQQQQMLAERELRSFFKLPSPISTRRSETEDGETKKETNVSAVLFKGPG 154
+GR+ +LL + ++ ++ RE F+ P S + E+ E + N+ ++FKGP
Sbjct 517 KGRIYQLLAKASYKKAIILTREATGHFQESEPFS--HIDPEESEETRLLNILGLIFKGPA 574
Query 155 PPT 157
T
Sbjct 575 AST 577
> At3g08850
Length=1384
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 31/62 (50%), Gaps = 5/62 (8%)
Query 66 GASISAAAAKALGAA----ALIPLQPPLYGWRCRGRLQELLQEQQQQQQMLAERELRSFF 121
G+ S A LGA+ +L+PL +YGW C + LL Q++ A+RE + F
Sbjct 980 GSPDSGLADPLLGASGSERSLLPLST-IYGWSCGHFSKPLLGGADASQEIAAKREEKEKF 1038
Query 122 KL 123
L
Sbjct 1039 AL 1040
Lambda K H
0.318 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2389760076
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40