bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2461_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
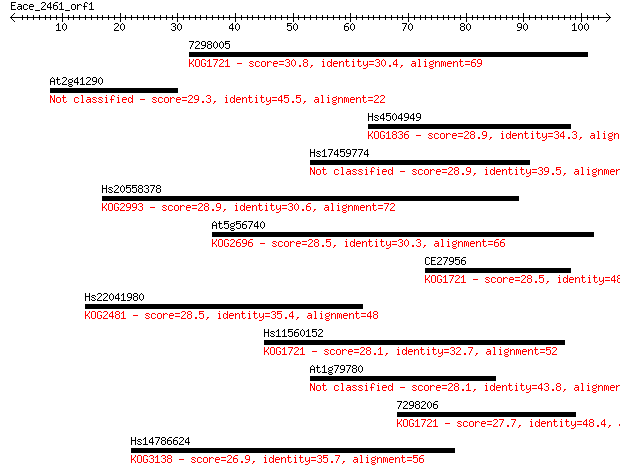
7298005 30.8 0.56
At2g41290 29.3 1.7
Hs4504949 28.9 2.1
Hs17459774 28.9 2.3
Hs20558378 28.9 2.5
At5g56740 28.5 2.8
CE27956 28.5 3.0
Hs22041980 28.5 3.1
Hs11560152 28.1 4.2
At1g79780 28.1 4.5
7298206 27.7 4.9
Hs14786624 26.9 8.1
> 7298005
Length=747
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 36/72 (50%), Gaps = 4/72 (5%)
Query 32 IQTRFLYERLSENKINSYLYPLSSRIPVEKHGFPPPMLATHHLRSHSPGEPNICSSVQGQ 91
I FL ++L + K N++++PLS V PP LA ++ + P + N C++
Sbjct 48 IGQDFLEQQLEQYKANNFMFPLSMAGFVSADSAPPGDLAKENMENSLP-DGNPCNNNNDD 106
Query 92 EF---KKRSNHA 100
E K R N++
Sbjct 107 ELPQCKIRRNYS 118
> At2g41290
Length=376
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 10/22 (45%), Positives = 11/22 (50%), Gaps = 0/22 (0%)
Query 8 CFEPHHHTHSHEVCSRPLNAGF 29
C PH H + VC RPL F
Sbjct 96 CEGPHEHQRTEHVCGRPLGLAF 117
> Hs4504949
Length=1816
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 63 GFPPPMLATHHLRSHSPGEPNICSSVQGQEFKKRS 97
G PP +L + LR+H P + N ++G +F+K+
Sbjct 1170 GAPPEILQSRALRAHLPLDINFRGCMKGFQFQKKD 1204
> Hs17459774
Length=211
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 53 LSSRIPVEKHGFPPPMLATHHLRSHSPGEPNICSSVQG 90
L+S++P+E G + HL S S G C+SVQG
Sbjct 118 LASQMPMETTGSQVAVANQPHLNSRSKGGHTYCNSVQG 155
> Hs20558378
Length=1938
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 17 SHEVCSRPLNAGFRSIQTRFLYERLSENKINSYLYP-LSSRIPVEKHGFPPPMLATHHLR 75
S E+ RPL+ +Q + +RL+ ++IN +LY S R+P H + A H
Sbjct 1508 SQELEERPLSRQVFIVQELEVRDRLASSQINKFLYLHTSERMPRRAHSNMLTIKALHVAP 1567
Query 76 SHSPGEPNICSSV 88
+ + G P C V
Sbjct 1568 TTNLGGPECCLRV 1580
> At5g56740
Length=467
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 0/66 (0%)
Query 36 FLYERLSENKINSYLYPLSSRIPVEKHGFPPPMLATHHLRSHSPGEPNICSSVQGQEFKK 95
+L E ++ I +Y L+ P EK + + LRS P +P+I S+VQ K
Sbjct 263 YLMEVVNNVAITENVYDLTVEEPSEKFQHIRTCIDINRLRSFDPIKPDIDSAVQTLTKGK 322
Query 96 RSNHAE 101
S A+
Sbjct 323 LSKKAQ 328
> CE27956
Length=982
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 73 HLRSHSPGEPNICSSVQGQEFKKRS 97
H+R H+PG P +CS Q Q F + S
Sbjct 947 HMRFHTPGNPFMCSDCQYQAFNELS 971
> Hs22041980
Length=568
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 14 HTHSHEVCSRPLNAGFRSIQTRFLYERLSENKINSYLYPLSSRIPVEK 61
H H +V RPL RS+ + + +E K + Y L S PVEK
Sbjct 278 HDHRTDVDYRPLPVAQRSLSPQAEGQAQAEGKASEGTYALDSESPVEK 325
> Hs11560152
Length=1342
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 31/53 (58%), Gaps = 9/53 (16%)
Query 45 KINSYLYPLSSRI-PVEKHGFPPPMLATHHLRSHSPGEPNICSSVQGQEFKKR 96
K++S+ P+ R+ P++K T H+++HS +P++C G+ FKKR
Sbjct 558 KLSSFPCPVCGRVYPMQKR-------LTQHMKTHSTEKPHMCDKC-GKSFKKR 602
> At1g79780
Length=168
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 53 LSSRIPVEKHGFPPPMLATHHLRSHSPGEPNI 84
L +R + H FP P LA HLRS G N+
Sbjct 85 LQTRHRISHHAFPLPSLALLHLRSAGSGVTNL 116
> 7298206
Length=593
Score = 27.7 bits (60), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 68 MLATHHLRSHSPGEPNICSSVQGQEFKKRSN 98
ML H+R+HS +P C SV G+ F RSN
Sbjct 430 MLLKQHMRTHSGEKPYQC-SVCGKSFADRSN 459
> Hs14786624
Length=782
Score = 26.9 bits (58), Expect = 8.1, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 8/64 (12%)
Query 22 SRPLNAGFRSIQTRFLYERLSENKI---NSYLYPLSSRI--PVEKHGF---PPPMLATHH 73
S P GFR T+FLY + + S + P +SRI P + + PP +
Sbjct 559 SLPSRKGFRHQTTKFLYRLVGSEDMAVDQSIVSPYTSRILKPYIRRDYETKPPKLQLLSQ 618
Query 74 LRSH 77
+RSH
Sbjct 619 IRSH 622
Lambda K H
0.317 0.131 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40