bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2470_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
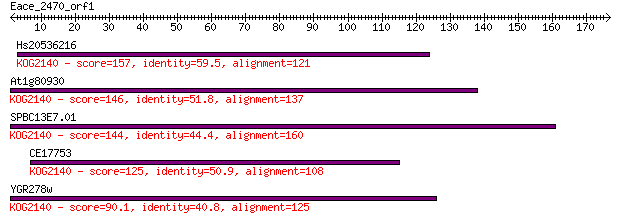
Hs20536216 157 1e-38
At1g80930 146 3e-35
SPBC13E7.01 144 1e-34
CE17753 125 4e-29
YGR278w 90.1 2e-18
> Hs20536216
Length=908
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 72/121 (59%), Positives = 86/121 (71%), Gaps = 0/121 (0%)
Query 3 FKRQYTLAHRLETAKLRNTAKFFAHLLTSDAVPWSVLEVFELTEEKTTSSGRIFLKVLLQ 62
FK QY HRLET KLRN AK FAHLL +D++PWSVLE +L+EE TTSS RIF+K+ Q
Sbjct 529 FKEQYDTIHRLETNKLRNVAKMFAHLLYTDSLPWSVLECIKLSEETTTSSSRIFVKIFFQ 588
Query 63 DTAETMGLKTLYERLMHPDLKPYVKGLFPEDNPRSLRFCINFFTAIGLGGLTDNQRQILA 122
+ E MGL L RL L+P+ +GL P DNPR+ RF INFFT+IGLGGLTD R+ L
Sbjct 589 ELCEYMGLPKLNARLKDETLQPFFEGLLPRDNPRNTRFAINFFTSIGLGGLTDELREHLK 648
Query 123 G 123
Sbjct 649 N 649
> At1g80930
Length=900
Score = 146 bits (368), Expect = 3e-35, Method: Composition-based stats.
Identities = 71/137 (51%), Positives = 102/137 (74%), Gaps = 2/137 (1%)
Query 1 EAFKRQYTLAHRLETAKLRNTAKFFAHLLTSDAVPWSVLEVFELTEEKTTSSGRIFLKVL 60
+ F +QY++ HRLET KLRN AKFFAHLL +DA+PW VL LTEE TTSS RIF+K+L
Sbjct 722 KCFVQQYSMIHRLETNKLRNVAKFFAHLLGTDALPWHVLAYIRLTEEDTTSSSRIFIKIL 781
Query 61 LQDTAETMGLKTLYERLMHPDLKPYVKGLFPEDNPRSLRFCINFFTAIGLGGLTDNQRQI 120
Q+ +E +G++ L ERL P ++ ++ +FP+DNP++ RF INFFT+IGLGG+T+N R+
Sbjct 782 FQELSEHLGIRLLNERLQDPTMQESLESIFPKDNPKNTRFAINFFTSIGLGGITENLREY 841
Query 121 LAGLQHQAHLQQQQQQL 137
L + + + Q+Q+Q+
Sbjct 842 LKNM--PSLIMQRQKQV 856
> SPBC13E7.01
Length=628
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 71/160 (44%), Positives = 95/160 (59%), Gaps = 15/160 (9%)
Query 1 EAFKRQYTLAHRLETAKLRNTAKFFAHLLTSDAVPWSVLEVFELTEEKTTSSGRIFLKVL 60
+ FK Y HR ET +LRN A FFA+LL++D++ W V + LTE+ TT+S RIFLK++
Sbjct 484 QCFKNYYETIHRYETNRLRNIALFFANLLSTDSIGWEVYDCVRLTEDDTTASSRIFLKIM 543
Query 61 LQDTAETMGLKTLYERLMHPDLKPYVKGLFPEDNPRSLRFCINFFTAIGLGGLTDNQRQI 120
Q+ E +GLK+L ERL P+L PY+ GLFP D R++RF IN+FT+IGLG LT+ R+
Sbjct 544 FQEIVEALGLKSLVERLHDPNLVPYLHGLFPVDEARNVRFSINYFTSIGLGALTEEMREY 603
Query 121 LAGLQHQAHLQQQQQQLRQPSSSSSSSSSSSSSSSSSSSS 160
L L P S S SS S + S
Sbjct 604 L---------------LTMPKSEPKEQDSEGYSSGSETGS 628
> CE17753
Length=897
Score = 125 bits (315), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 55/108 (50%), Positives = 72/108 (66%), Gaps = 0/108 (0%)
Query 7 YTLAHRLETAKLRNTAKFFAHLLTSDAVPWSVLEVFELTEEKTTSSGRIFLKVLLQDTAE 66
Y+ HR++ KLRN A+ AHLL++DA+ W +L ++TEE TTSSGRI++K + + E
Sbjct 564 YSTIHRIDITKLRNLARLIAHLLSTDAIDWKILADMKMTEEDTTSSGRIYIKYIFNELVE 623
Query 67 TMGLKTLYERLMHPDLKPYVKGLFPEDNPRSLRFCINFFTAIGLGGLT 114
MG+ L+ R+ P L GLFP NP S RF INFFT IGLGGLT
Sbjct 624 AMGMVKLHSRVTDPTLAHCFVGLFPRTNPNSARFSINFFTMIGLGGLT 671
> YGR278w
Length=577
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 51/127 (40%), Positives = 74/127 (58%), Gaps = 6/127 (4%)
Query 1 EAFKRQYTL-AHRLETAKLRNTAKFFAHLLTSDAVPWSVLEVFELTEEKTTSSGRIFLKV 59
E F++ YT ET +LR KF+ HL++ + +P L++ +LTEE++ GRIF+K
Sbjct 363 ETFEQNYTQDIEDYETDQLRILGKFWGHLISYEFLPMDCLKIIKLTEEESCPQGRIFIKF 422
Query 60 LLQDTAETMGLKTLYERLMHPDLKPYVKGLFP-EDNPRSLRFCINFFTAIGLGGLTDNQR 118
L Q+ +GL L RL L G+FP E + +R+ INFFTAIGLG LT++ R
Sbjct 423 LFQELVNELGLDELQLRLNSSKL----DGMFPLEGDAEHIRYSINFFTAIGLGLLTEDMR 478
Query 119 QILAGLQ 125
L +Q
Sbjct 479 SRLTIIQ 485
Lambda K H
0.310 0.121 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2707167450
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40