bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2478_orf2
Length=161
Score E
Sequences producing significant alignments: (Bits) Value
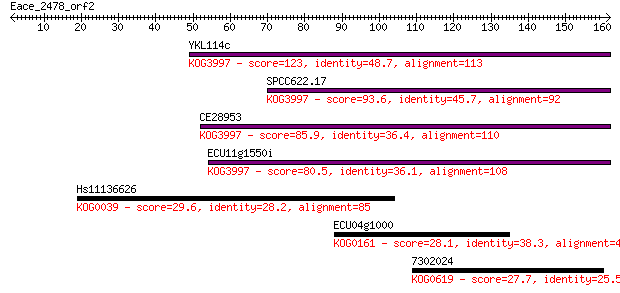
YKL114c 123 2e-28
SPCC622.17 93.6 2e-19
CE28953 85.9 3e-17
ECU11g1550i 80.5 1e-15
Hs11136626 29.6 2.9
ECU04g1000 28.1 8.1
7302024 27.7 9.8
> YKL114c
Length=367
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 55/113 (48%), Positives = 75/113 (66%), Gaps = 0/113 (0%)
Query 49 AAKSRKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERC 108
+A S+ GAH+S AGG+ N++ + FN +FA+FLK RKWVS T+ I+ FK+ C
Sbjct 10 SAVSKYKFGAHMSGAGGISNSVTNAFNTGCNSFAMFLKSPRKWVSPQYTQEEIDKFKKNC 69
Query 109 DQLEMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
+ T VLPHG Y IN+ANPD+ K E +Y++ +DDL RCE LGI LYN+
Sbjct 70 ATYNYNPLTDVLPHGQYFINLANPDREKAEKSYESFMDDLNRCEQLGIGLYNL 122
> SPCC622.17
Length=366
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 42/92 (45%), Positives = 59/92 (64%), Gaps = 0/92 (0%)
Query 70 LISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQLEMDMATQVLPHGSYLINV 129
L++ F + + A F+K QRKW S L+E F E +++ D + QVL HGSYLIN+
Sbjct 9 LLTKFYISANSCAFFVKSQRKWTSPDLSEDVAQKFLETASEMKFDASKQVLVHGSYLINM 68
Query 130 ANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
AN D+ K+E A+ +DDL+RCE LG+ LYN
Sbjct 69 ANADEQKREQAFNCFVDDLKRCERLGVGLYNF 100
> CE28953
Length=396
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 40/110 (36%), Positives = 66/110 (60%), Gaps = 1/110 (0%)
Query 52 SRKFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQL 111
S K +G H+SAAGG+ A+ + ++FA+F++ QR W P++E + + + +
Sbjct 116 SSKMLGFHVSAAGGLEQAIYNARAEGCRSFAMFVRNQRTWNHKPMSEEVVENWWKAVRET 175
Query 112 EMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
+ Q++PHGSYL+N +P+ K E + A+LD+ QR E LGI +YN
Sbjct 176 NFPL-DQIVPHGSYLMNAGSPEAEKLEKSRLAMLDECQRAEKLGITMYNF 224
> ECU11g1550i
Length=275
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 64/108 (59%), Gaps = 7/108 (6%)
Query 54 KFVGAHISAAGGVHNALISCFNVKGQAFALFLKCQRKWVSSPLTETAINGFKERCDQLEM 113
K +GAH+S + G+H + A+FLK QR++++ P+ + A F+ E+
Sbjct 3 KLIGAHLSISKGIHTIQAQMELLGLDTCAIFLKSQRRYLTPPMKQGAAEMFRAHVRHPEL 62
Query 114 DMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLYNI 161
++PHGSYLIN NPD + + + L+DDL+RC++LGI +YN+
Sbjct 63 -----IVPHGSYLINFGNPDLV--DKSMECLVDDLERCKSLGIGMYNM 103
> Hs11136626
Length=568
Score = 29.6 bits (65), Expect = 2.9, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 31/85 (36%), Gaps = 9/85 (10%)
Query 19 KAGKKHATATTAPQSSPLQPDEEFNALAALAAKSRKFVGAHISAAGGVHNALISCFNVKG 78
K G K A P E++ +A F HI AAG AL+ F +G
Sbjct 312 KRGFKMAPGQYILVQCPAISSLEWHPFTLTSAPQEDFFSVHIRAAGDWTAALLEAFGAEG 371
Query 79 QAFALFLKCQRKWVSSPLTETAING 103
QA Q W L A++G
Sbjct 372 QAL------QEPW---SLPRLAVDG 387
> ECU04g1000
Length=1700
Score = 28.1 bits (61), Expect = 8.1, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 88 QRKWVSSPLTETA-INGF--KERCDQLEMDMATQVLPHGSYLINVANPDK 134
++KWV +P ++ A + GF KE D LE+D ++ H S + NP K
Sbjct 53 EKKWVWAPSSKEAYVCGFVVKEEGDVLEIDCRGVIVRHKSCEVFRMNPPK 102
> 7302024
Length=1315
Score = 27.7 bits (60), Expect = 9.8, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 109 DQLEMDMATQVLPHGSYLINVANPDKTKQENAYKALLDDLQRCETLGIKLY 159
D + + TQ LPH L+ NP + N++ + +DL+ + +L+
Sbjct 983 DLTNVPLMTQALPHLRRLMLSGNPITSLNNNSFDGVNEDLEMLDISNFRLH 1033
Lambda K H
0.316 0.130 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2208717646
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40