bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2503_orf1
Length=101
Score E
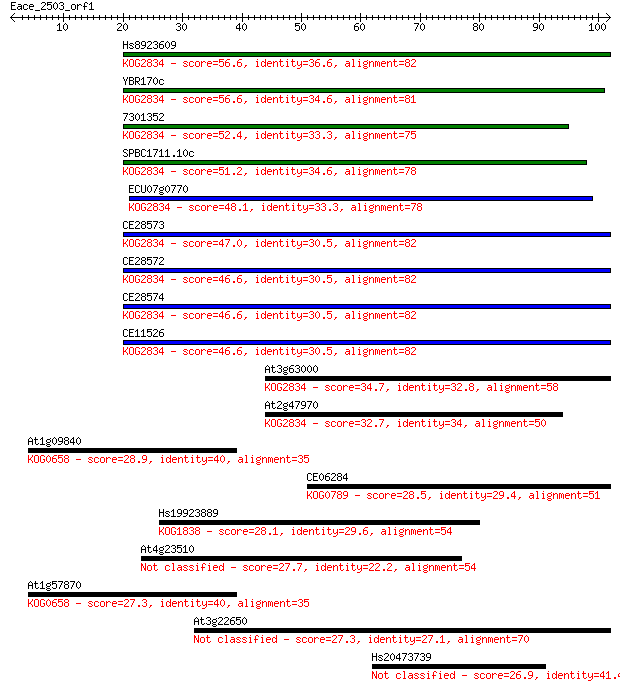
Sequences producing significant alignments: (Bits) Value
Hs8923609 56.6 1e-08
YBR170c 56.6 1e-08
7301352 52.4 2e-07
SPBC1711.10c 51.2 4e-07
ECU07g0770 48.1 3e-06
CE28573 47.0 1e-05
CE28572 46.6 1e-05
CE28574 46.6 1e-05
CE11526 46.6 1e-05
At3g63000 34.7 0.049
At2g47970 32.7 0.17
At1g09840 28.9 2.4
CE06284 28.5 3.0
Hs19923889 28.1 4.5
At4g23510 27.7 5.4
At1g57870 27.3 6.5
At3g22650 27.3 8.0
Hs20473739 26.9 8.7
> Hs8923609
Length=608
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 40/82 (48%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L Q YR VD++ N A F+ FWR + Q+FG ++G YTE G
Sbjct 210 PSAITLNRQKYRHVDNIMFENHTVADRFLDFWRKT---GNQQFGYLYGRYTEHKDIPLGI 266
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
R IYEPPQ G L+
Sbjct 267 RAEVAAIYEPPQIGTQNSLELL 288
> YBR170c
Length=580
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L+ Q +R VDH+E +F+ WR + QRFG M+G Y++ ++ G
Sbjct 221 PSAITLQQQEFRMVDHVEFQKSEIINEFIQAWRYT---GMQRFGYMYGSYSKYDNTPLGI 277
Query 80 RPVCEVIYEPPQEGVGEGWRL 100
+ V E IYEPPQ +G +
Sbjct 278 KAVVEAIYEPPQHDEQDGLTM 298
> 7301352
Length=1006
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P ++L Q YR VD++ N + F+++WR + QR G ++G Y + G
Sbjct 244 PSAITLNRQTYRHVDNVMFENTKIVERFLNYWRTT---GHQRMGYLYGTYEQHTDVPLGI 300
Query 80 RPVCEVIYEPPQEGV 94
R IYEPPQE
Sbjct 301 RAKVAAIYEPPQEST 315
> SPBC1711.10c
Length=545
Score = 51.2 bits (121), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/78 (34%), Positives = 41/78 (52%), Gaps = 3/78 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
P T+ L Q +R +DH+E + F++ WR S QR G +G++ + N+ G
Sbjct 226 PSTVMLNLQPFRVIDHIEFASPGIVDSFLNKWRQS---GFQRIGYTYGHFEQYNNVPLGI 282
Query 80 RPVCEVIYEPPQEGVGEG 97
+ V E IYEPPQ +G
Sbjct 283 KGVIEAIYEPPQVSEADG 300
> ECU07g0770
Length=505
Score = 48.1 bits (113), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 41/78 (52%), Gaps = 3/78 (3%)
Query 21 QTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGSR 80
+ + L QV+R VDH+E +F+ WR+S +QRFG + G Y + G++
Sbjct 165 KNIILAPQVFRMVDHVEFDGKHLVENFIRNWRES---GRQRFGFLVGRYMDHEMIPLGTK 221
Query 81 PVCEVIYEPPQEGVGEGW 98
V I+EP QE +G+
Sbjct 222 AVVSGIWEPEQEDYPDGF 239
> CE28573
Length=529
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
PQ ++L Q +R VD++++ N F+ +WR S QR G + G Y G
Sbjct 113 PQVVTLNRQKFRHVDNIQIENQELVNQFLDYWRLS---GHQRVGFLIGQYQPHLEVPLGI 169
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
+ IYEPPQ +G +
Sbjct 170 KATVAAIYEPPQHCREDGIEFL 191
> CE28572
Length=527
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
PQ ++L Q +R VD++++ N F+ +WR S QR G + G Y G
Sbjct 113 PQVVTLNRQKFRHVDNIQIENQELVNQFLDYWRLS---GHQRVGFLIGQYQPHLEVPLGI 169
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
+ IYEPPQ +G +
Sbjct 170 KATVAAIYEPPQHCREDGIEFL 191
> CE28574
Length=529
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
PQ ++L Q +R VD++++ N F+ +WR S QR G + G Y G
Sbjct 113 PQVVTLNRQKFRHVDNIQIENQELVNQFLDYWRLS---GHQRVGFLIGQYQPHLEVPLGI 169
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
+ IYEPPQ +G +
Sbjct 170 KATVAAIYEPPQHCREDGIEFL 191
> CE11526
Length=527
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 3/82 (3%)
Query 20 PQTLSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGS 79
PQ ++L Q +R VD++++ N F+ +WR S QR G + G Y G
Sbjct 113 PQVVTLNRQKFRHVDNIQIENQELVNQFLDYWRLS---GHQRVGFLIGQYQPHLEVPLGI 169
Query 80 RPVCEVIYEPPQEGVGEGWRLI 101
+ IYEPPQ +G +
Sbjct 170 KATVAAIYEPPQHCREDGIEFL 191
> At3g63000
Length=413
Score = 34.7 bits (78), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 27/58 (46%), Gaps = 6/58 (10%)
Query 44 ARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQEGVGEGWRLI 101
A F F +SL +R G M+G +ED IYEPPQ+G+ + L+
Sbjct 139 ANAFQHFVNESLAFAVKRGGFMYGNVSEDGQVE------VNFIYEPPQQGMEDNLILM 190
> At2g47970
Length=411
Score = 32.7 bits (73), Expect = 0.17, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 6/50 (12%)
Query 44 ARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGSRPVCEVIYEPPQEG 93
A F + +SL +R G M+G TE+ + IYEPPQ+G
Sbjct 137 ANAFQHYVNESLAFAVKRGGFMYGTVTEEGQVE------VDFIYEPPQQG 180
> At1g09840
Length=421
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 4 KSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLEL 38
K YK +LQ QM P ++LKH + R D+ E+
Sbjct 118 KRYKNRELQIMQMLDHPNAVALKHSFFSRTDNEEV 152
> CE06284
Length=621
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Query 51 WRDSLCMQQQRFGLMFGYYTEDNHYN--KGSRPVCEVIYEPPQEGVGEGWRLI 101
++D +C + R L GY + H N KG++P+ + P ++ V + WR++
Sbjct 263 YKDVMCNDKTRVILKDGYPGDYIHANYLKGTKPMLILTQGPLKDSVMDIWRMV 315
> Hs19923889
Length=409
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 26/55 (47%), Gaps = 12/55 (21%)
Query 26 KHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRF-GLMFGYYTEDNHYNKGS 79
+H ++VD +M + R+F +RF +MFGY T D++Y S
Sbjct 283 RHMFVKQVDMDHVMKAKSIREF-----------DKRFTSVMFGYQTIDDYYTDAS 326
> At4g23510
Length=867
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 12/54 (22%), Positives = 25/54 (46%), Gaps = 0/54 (0%)
Query 23 LSLKHQVYRRVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYN 76
L + V ++VD + ++ E D + WR +L R G + + +++ N
Sbjct 354 LKTEEDVRKKVDRSNIRSILETEDMIWGWRQALVSVGGRMGFSYNHKCDNDFVN 407
> At1g57870
Length=420
Score = 27.3 bits (59), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 4 KSYKPTKLQAGQMNRMPQTLSLKHQVYRRVDHLEL 38
K YK +LQ QM P + LKH Y R ++ E+
Sbjct 117 KRYKNRELQIMQMLDHPNVVCLKHSFYSRTENEEV 151
> At3g22650
Length=372
Score = 27.3 bits (59), Expect = 8.0, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 31/73 (42%), Gaps = 10/73 (13%)
Query 32 RVDHLELMNVREARDFVSFWRDSLCMQQQRFGLMFGYYTEDNHYNKGSRPVCEVIYEPP- 90
RV +E M+ + DF F D++C + + +F ED GS YEP
Sbjct 124 RVKWIEPMDFYSSNDFYGFGYDNVCRDEYKLLRIFDGEIEDESEIAGS-------YEPKI 176
Query 91 --QEGVGEGWRLI 101
+ + WR++
Sbjct 177 QIYDFKSDSWRIV 189
> Hs20473739
Length=565
Score = 26.9 bits (58), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 15/29 (51%), Gaps = 0/29 (0%)
Query 62 FGLMFGYYTEDNHYNKGSRPVCEVIYEPP 90
F FG YT++ H +GS E EPP
Sbjct 441 FTYPFGPYTKEKHQPRGSNTTTEFNTEPP 469
Lambda K H
0.323 0.138 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40