bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2546_orf1
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
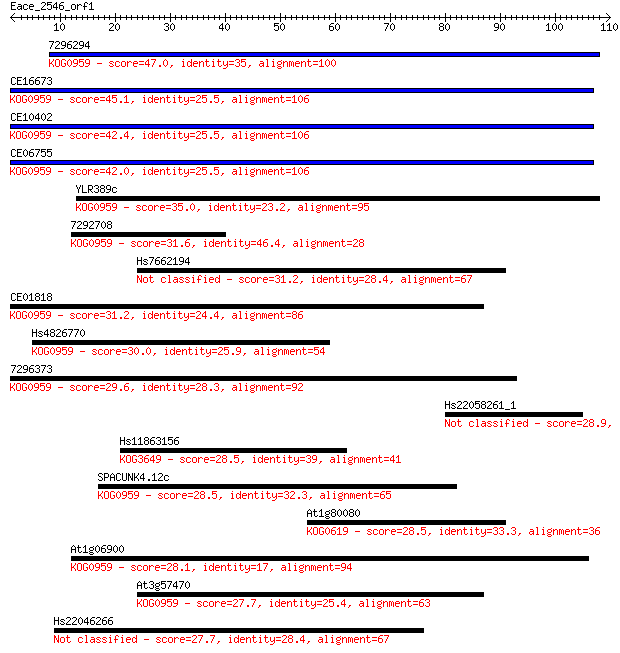
7296294 47.0 9e-06
CE16673 45.1 3e-05
CE10402 42.4 2e-04
CE06755 42.0 3e-04
YLR389c 35.0 0.034
7292708 31.6 0.39
Hs7662194 31.2 0.51
CE01818 31.2 0.51
Hs4826770 30.0 1.0
7296373 29.6 1.5
Hs22058261_1 28.9 2.6
Hs11863156 28.5 3.0
SPACUNK4.12c 28.5 3.1
At1g80080 28.5 3.6
At1g06900 28.1 4.4
At3g57470 27.7 5.2
Hs22046266 27.7 5.6
> 7296294
Length=990
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 51/103 (49%), Gaps = 4/103 (3%)
Query 8 DRVNLAV--LTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVV 65
D N+ V ++Q L+ YD LRT+ QLGYI + + ++ V+ AK HP V
Sbjct 776 DHTNIMVNLVSQVLSEPCYDCLRTKEQLGYIVFSGVRKVNGANGIRIIVQSAK-HPSYVE 834
Query 66 KMIDEELTKAKEYLANMPDAEMARWKEA-AHAKLTKREATFSE 107
I+ L + + +MP E R KEA A KL K + F +
Sbjct 835 DRIENFLQTYLQVIEDMPLDEFERHKEALAVKKLEKPKTIFQQ 877
> CE16673
Length=301
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 49/106 (46%), Gaps = 1/106 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ + +D+LRT+ LGYI + T LQ V+G K+
Sbjct 80 IGVQNTYDNAVIGLIKNLITEPAFDTLRTKESLGYIVWTRTHFNCGTVALQILVQGPKS- 138
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKREATFS 106
D V++ I+ L ++ + MP E A+L ++ T S
Sbjct 139 VDHVLERIEAFLESVRKEIVEMPQEEFENRVSGLIAQLEEKPKTLS 184
> CE10402
Length=1067
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 48/106 (45%), Gaps = 1/106 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ Q + +++LRT LGYI T L V+G K+
Sbjct 839 IGVQNTYDNAVVGLIDQLIREPAFNTLRTNEALGYIVWTGSRLNCGTVALNVIVQGPKS- 897
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKREATFS 106
D V++ I+ L ++ +A MP E A+L ++ T S
Sbjct 898 VDHVLERIEVFLESVRKEIAEMPQEEFDNQVSGMIARLEEKPKTLS 943
> CE06755
Length=980
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/106 (25%), Positives = 49/106 (46%), Gaps = 1/106 (0%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ + D + ++ Q + ++D+LRT LGYI L FV+G K+
Sbjct 753 IGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEALGYIVWTGCRFNCGAVALNIFVQGPKS- 811
Query 61 PDEVVKMIDEELTKAKEYLANMPDAEMARWKEAAHAKLTKREATFS 106
D V++ I+ L ++ + MP E + A+L ++ T S
Sbjct 812 VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAGMIARLEEKPKTLS 857
> YLR389c
Length=988
Score = 35.0 bits (79), Expect = 0.034, Method: Composition-based stats.
Identities = 22/95 (23%), Positives = 44/95 (46%), Gaps = 1/95 (1%)
Query 13 AVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEEL 72
+ Q ++ +D+LRT+ QLGY+ + TA ++ ++ T P + I+
Sbjct 818 GLFAQLIHEPCFDTLRTKEQLGYVVFSSSLNNHGTANIRILIQSEHTTP-YLEWRINNFY 876
Query 73 TKAKEYLANMPDAEMARWKEAAHAKLTKREATFSE 107
+ L +MP+ + + KEA L ++ +E
Sbjct 877 ETFGQVLRDMPEEDFEKHKEALCNSLLQKFKNMAE 911
> 7292708
Length=1077
Score = 31.6 bits (70), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 12 LAVLTQFLNRRIYDSLRTEAQLGYIAGA 39
L +L F++ ++D LRT+ QLGY GA
Sbjct 827 LDLLMMFVDEPLFDQLRTKEQLGYHVGA 854
> Hs7662194
Length=1462
Score = 31.2 bits (69), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query 24 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLANMP 83
+D + + AG E+ A + L + E + TH EV +M+ E+L + Y+ M
Sbjct 895 HDRVHLRSTYHRYAGHLEASADCSRALS-YYEKSDTHRFEVPRMLSEDLPSLELYVNKMK 953
Query 84 DAEMARW 90
D + RW
Sbjct 954 DKTLWRW 960
> CE01818
Length=745
Score = 31.2 bits (69), Expect = 0.51, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 37/86 (43%), Gaps = 1/86 (1%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH 60
+G+ S + +L + + Y LRT LGY + L V+G ++
Sbjct 587 IGVQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVSTESRLNDGNVYLHVIVQGPES- 645
Query 61 PDEVVKMIDEELTKAKEYLANMPDAE 86
D V++ I+ L A+E + MP +
Sbjct 646 ADHVLERIEVFLESAREEIVAMPQED 671
> Hs4826770
Length=1019
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 5 SIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAK 58
S + + L + Q ++ +++LRT+ QLGYI + +A L+ ++ K
Sbjct 801 STSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANGIQSLRFIIQSEK 854
> 7296373
Length=908
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 26/94 (27%), Positives = 39/94 (41%), Gaps = 7/94 (7%)
Query 1 LGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKE--SQAASTALLQCFVEGAK 58
L + I D V L V F N+ LRT+ QLGY G + L+ + K
Sbjct 667 LKMECIMDLVELIVEEPFFNQ-----LRTQEQLGYSLGIHQRIGYGVLAFLITINTQETK 721
Query 59 THPDEVVKMIDEELTKAKEYLANMPDAEMARWKE 92
D V + I+ ++ E ++ M D E +E
Sbjct 722 HRADYVEQRIEAFRSRMAELVSQMSDTEFKNIRE 755
> Hs22058261_1
Length=354
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 1/26 (3%)
Query 80 ANMPDAEMARWKEA-AHAKLTKREAT 104
A MPD E+ARWK+ AK ++AT
Sbjct 283 AEMPDQEIARWKQRLEQAKAEMKQAT 308
> Hs11863156
Length=971
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query 21 RRIYDSLRTEAQL--GYIAGAKESQAASTALLQCFVEGAKT-HP 61
+RI S +TE ++ G I G K L QCF+E +++ HP
Sbjct 230 KRILMSKKTEMEIVDGLIEGCKTQPLPQDPLWQCFLESSQSVHP 273
> SPACUNK4.12c
Length=969
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 21/79 (26%), Positives = 31/79 (39%), Gaps = 14/79 (17%)
Query 17 QFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTH--------------PD 62
Q + + LRT+ QLGYI Q L FV+ ++
Sbjct 774 QIMKEPTFSILRTKEQLGYIVFTLVRQVTPFINLNIFVQSERSSTYLESRIRALLDQFKS 833
Query 63 EVVKMIDEELTKAKEYLAN 81
E ++M DE+ +K K L N
Sbjct 834 EFLEMSDEDFSKHKSSLIN 852
> At1g80080
Length=496
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 55 EGAKTHPDEVVKMIDEELTKAKEYLANMPDAEMARW 90
+GA+T PDE + D ++ A +PD RW
Sbjct 50 DGARTEPDEQDAVYDIMRATGNDWAAAIPDVCRGRW 85
> At1g06900
Length=1023
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 16/94 (17%), Positives = 39/94 (41%), Gaps = 0/94 (0%)
Query 12 LAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEE 71
L + + + +++ LRT+ QLGY+ V+ +K P ++ +D
Sbjct 835 LDLFHEIIEEPLFNQLRTKEQLGYVVECGPRLTYRVHGFCFCVQSSKYGPVHLLGRVDNF 894
Query 72 LTKAKEYLANMPDAEMARWKEAAHAKLTKREATF 105
+ + L + D ++ A+L +++ +
Sbjct 895 IKDIEGLLEQLDDESYEDYRSGMIARLLEKDPSL 928
> At3g57470
Length=989
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 16/63 (25%), Positives = 26/63 (41%), Gaps = 0/63 (0%)
Query 24 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMIDEELTKAKEYLANMP 83
+ LRT QLGYI S + +Q ++ + P + ++ L + NM
Sbjct 809 FHQLRTIEQLGYITSLSLSNDSGVYGVQFIIQSSVKGPGHIDSRVESLLKDLESKFYNMS 868
Query 84 DAE 86
D E
Sbjct 869 DEE 871
> Hs22046266
Length=390
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 24/67 (35%), Gaps = 0/67 (0%)
Query 9 RVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKTHPDEVVKMI 68
R V + L R I LR A K LL CFV+ HP + +
Sbjct 317 RPQCRVKVKLLQRSISSLLRFAAGEDGSYEVKSVLGKEVGLLNCFVQSVTAHPTSCIGLE 376
Query 69 DEELTKA 75
+ EL A
Sbjct 377 EIELLSA 383
Lambda K H
0.315 0.128 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1199474506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40