bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
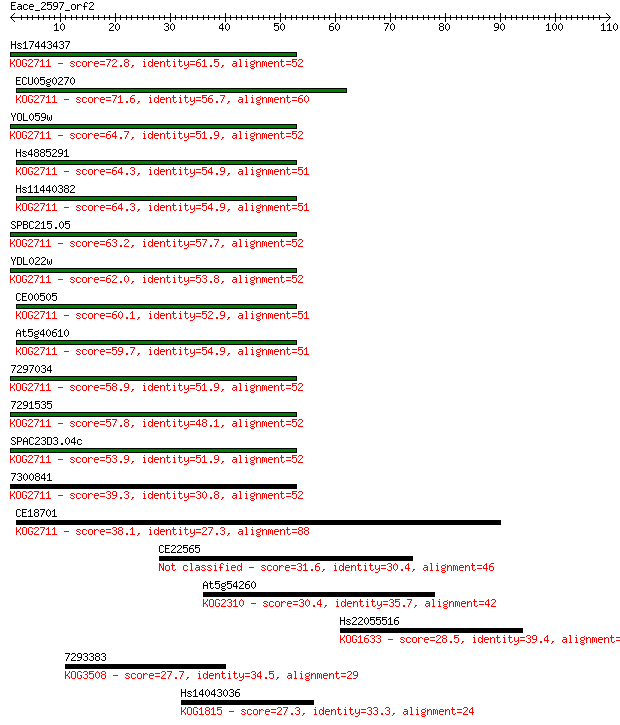
Query= Eace_2597_orf2
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
Hs17443437 72.8 2e-13
ECU05g0270 71.6 3e-13
YOL059w 64.7 4e-11
Hs4885291 64.3 5e-11
Hs11440382 64.3 5e-11
SPBC215.05 63.2 1e-10
YDL022w 62.0 3e-10
CE00505 60.1 1e-09
At5g40610 59.7 1e-09
7297034 58.9 2e-09
7291535 57.8 5e-09
SPAC23D3.04c 53.9 7e-08
7300841 39.3 0.002
CE18701 38.1 0.004
CE22565 31.6 0.41
At5g54260 30.4 0.80
Hs22055516 28.5 3.1
7293383 27.7 6.1
Hs14043036 27.3 6.6
> Hs17443437
Length=351
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 32/52 (61%), Positives = 39/52 (75%), Gaps = 0/52 (0%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEVEGRKLTDWIN 52
LKV +VGSGNWG+AV KI+ +N K F + V+MWV+EE V GRKLTD IN
Sbjct 6 LKVCIVGSGNWGSAVAKIIGNNVKKLQKFASTVKMWVFEETVNGRKLTDIIN 57
> ECU05g0270
Length=345
Score = 71.6 bits (174), Expect = 3e-13, Method: Composition-based stats.
Identities = 34/63 (53%), Positives = 45/63 (71%), Gaps = 3/63 (4%)
Query 2 KVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEVEGRKLTDWIN---IHPSWQ 58
KVS++G+GNWGTA+G+++A+N S IF+ DVRMW EE EGR L D IN I+P +
Sbjct 4 KVSIIGNGNWGTAMGRLLANNTVESTIFDKDVRMWGCREEYEGRFLNDIINSDRINPRYL 63
Query 59 SGV 61
GV
Sbjct 64 PGV 66
> YOL059w
Length=440
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 27/53 (50%), Positives = 40/53 (75%), Gaps = 1/53 (1%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAK-NSHIFNTDVRMWVYEEEVEGRKLTDWIN 52
KV+++GSGNWGT + K++A N + +SHIF +VRMWV++E++ LTD IN
Sbjct 84 FKVTVIGSGNWGTTIAKVIAENTELHSHIFEPEVRMWVFDEKIGDENLTDIIN 136
> Hs4885291
Length=349
Score = 64.3 bits (155), Expect = 5e-11, Method: Composition-based stats.
Identities = 28/51 (54%), Positives = 37/51 (72%), Gaps = 0/51 (0%)
Query 2 KVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEVEGRKLTDWIN 52
KV +VGSGNWG+A+ KIV NA F+ V MWV+EE++ G+KLT+ IN
Sbjct 5 KVCIVGSGNWGSAIAKIVGGNAAQLAQFDPRVTMWVFEEDIGGKKLTEIIN 55
> Hs11440382
Length=349
Score = 64.3 bits (155), Expect = 5e-11, Method: Composition-based stats.
Identities = 28/51 (54%), Positives = 37/51 (72%), Gaps = 0/51 (0%)
Query 2 KVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEVEGRKLTDWIN 52
KV +VGSGNWG+A+ KIV NA F+ V MWV+EE++ G+KLT+ IN
Sbjct 5 KVCIVGSGNWGSAIAKIVGGNAAQLAQFDPRVTMWVFEEDIGGKKLTEIIN 55
> SPBC215.05
Length=385
Score = 63.2 bits (152), Expect = 1e-10, Method: Composition-based stats.
Identities = 30/57 (52%), Positives = 39/57 (68%), Gaps = 5/57 (8%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAK-NSHIFNTDVRMWVYEEEV----EGRKLTDWIN 52
L + +VGSGNWGTA+ KI NA+ + H F + VRMWV+EEE+ E RKLT+ N
Sbjct 23 LSIGVVGSGNWGTAIAKICGENARAHGHHFRSKVRMWVFEEEIEYKGEKRKLTEVFN 79
> YDL022w
Length=391
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 28/53 (52%), Positives = 38/53 (71%), Gaps = 1/53 (1%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAKN-SHIFNTDVRMWVYEEEVEGRKLTDWIN 52
KV+++GSGNWGT + K+VA N K +F V+MWV+EEE+ G KLT+ IN
Sbjct 35 FKVTVIGSGNWGTTIAKVVAENCKGYPEVFAPIVQMWVFEEEINGEKLTEIIN 87
> CE00505
Length=351
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 27/52 (51%), Positives = 38/52 (73%), Gaps = 1/52 (1%)
Query 2 KVSLVGSGNWGTAVGKIVASNAKN-SHIFNTDVRMWVYEEEVEGRKLTDWIN 52
KV+++GSGNWG+A+ +IV S K+ F+ VRMWV+EE V G KL++ IN
Sbjct 5 KVTIIGSGNWGSAIARIVGSTTKSFPDEFDPTVRMWVFEEIVNGEKLSEVIN 56
> At5g40610
Length=400
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 28/52 (53%), Positives = 37/52 (71%), Gaps = 1/52 (1%)
Query 2 KVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEV-EGRKLTDWIN 52
KV++VGSGNWG+ K++ASNA F+ +VRMWV+EE + G KL D IN
Sbjct 56 KVTVVGSGNWGSVAAKLIASNALKLPSFHDEVRMWVFEEVLPNGEKLNDVIN 107
> 7297034
Length=360
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 27/52 (51%), Positives = 37/52 (71%), Gaps = 0/52 (0%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEVEGRKLTDWIN 52
+ V +VGSGNWG+A+ KIV +NA F V M+VYEE ++G+KLT+ IN
Sbjct 5 VNVCIVGSGNWGSAIAKIVGANAAALPEFEERVTMFVYEELIDGKKLTEIIN 56
> 7291535
Length=349
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 25/52 (48%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEVEGRKLTDWIN 52
+ + ++GSGNW T + + V N NS + V M+VYEE VEGRKLT+ IN
Sbjct 4 IMICIIGSGNWATTIARNVGRNVLNSQTLDEKVPMYVYEEIVEGRKLTEIIN 55
> SPAC23D3.04c
Length=373
Score = 53.9 bits (128), Expect = 7e-08, Method: Composition-based stats.
Identities = 27/57 (47%), Positives = 39/57 (68%), Gaps = 5/57 (8%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAK-NSHIFNTDVRMWVYEEEV--EGRK--LTDWIN 52
+ V ++GSGNWGTA+ KI NAK + IF+ V MW+YEE++ EG++ LT+ N
Sbjct 25 ISVGIIGSGNWGTAIAKICGENAKAHPDIFHPQVHMWMYEEKIQHEGKECNLTEVFN 81
> 7300841
Length=1118
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 33/52 (63%), Gaps = 1/52 (1%)
Query 1 LKVSLVGSGNWGTAVGKIVASNAKNSHIFNTDVRMWVYEEEVEGRKLTDWIN 52
LK+ ++G+ WG+A+ +V++N F++ V ++VY+E + L++ IN
Sbjct 5 LKICIIGAEGWGSAIAAVVSNNVLEGD-FDSRVHLYVYDEMIRDTALSEIIN 55
> CE18701
Length=374
Score = 38.1 bits (87), Expect = 0.004, Method: Composition-based stats.
Identities = 24/89 (26%), Positives = 42/89 (47%), Gaps = 16/89 (17%)
Query 2 KVSLVGSGNWGTAVGKIVASNAK-NSHIFNTDVRMWVYEEEVEGRKLTDWINIHPSWQSG 60
K+++VG GNWG+A+ +V K +F V +W + G ++ PS
Sbjct 23 KIAIVGGGNWGSAIACVVGKTVKAQDEVFQPIVSIWCRDSRKPG-------DLSPSIAET 75
Query 61 VVPTPTASDSAHLGPRWHLQGRRVWNAVL 89
+ +S H P++ L GRR+ + V+
Sbjct 76 I-------NSTHENPKY-LPGRRIPDNVV 96
> CE22565
Length=309
Score = 31.6 bits (70), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 28 IFNTDVRMWVYEEEVEGRKLTDWINIHPSWQSGVVPTPTASDSAHL 73
IFN++ W + + G + TD+INI + + VV T + ++L
Sbjct 181 IFNSNYMSWFFNPMLPGHEATDYINIAHTINNCVVSLATTAIYSYL 226
> At5g54260
Length=720
Score = 30.4 bits (67), Expect = 0.80, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 36 WVYEEEVEGRKLTDWINIHPSWQSGVVPTPTASDSAHLGPRW 77
W+ E EG ++DW NI Q+ V P + S H PR+
Sbjct 201 WMRPEVQEGCDVSDWFNILVLHQNRVKSNPKNAISEHFLPRF 242
> Hs22055516
Length=1060
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 61 VVPTPTASDSAHLGPRWHLQGRRVWNAVLLFCL 93
V+P P D+ +L R LQGR +V ++CL
Sbjct 13 VLPPPAPLDTTNLAGRRTLQGRAKMASVPVYCL 45
> 7293383
Length=1436
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 11 WGTAVGKIVASNAKNSHIFNTDVRMWVYE 39
W ++ K +A NA+++ + ++MWVYE
Sbjct 138 WIYSLRKSIAPNAEHTRRTDNSLKMWVYE 166
> Hs14043036
Length=500
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 8/24 (33%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 32 DVRMWVYEEEVEGRKLTDWINIHP 55
D+R+WV E+ + +T W+ + P
Sbjct 44 DIRLWVSVEDAQMHTVTIWLTVRP 67
Lambda K H
0.321 0.134 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1199474506
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40