bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2622_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
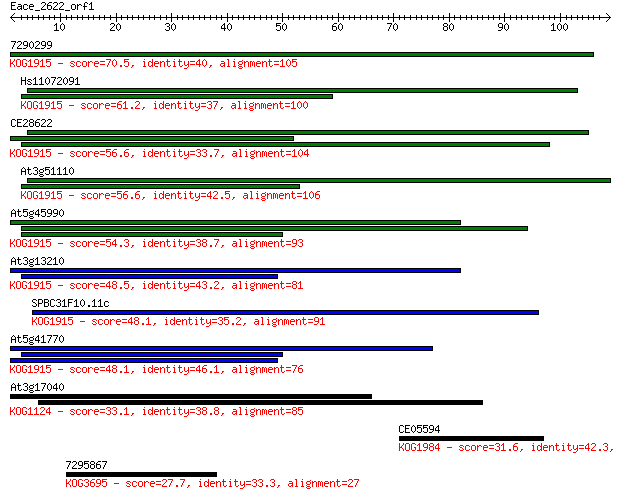
7290299 70.5 7e-13
Hs11072091 61.2 5e-10
CE28622 56.6 1e-08
At3g51110 56.6 1e-08
At5g45990 54.3 5e-08
At3g13210 48.5 3e-06
SPBC31F10.11c 48.1 4e-06
At5g41770 48.1 4e-06
At3g17040 33.1 0.12
CE05594 31.6 0.36
7295867 27.7 4.7
> 7290299
Length=702
Score = 70.5 bits (171), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 42/114 (36%), Positives = 62/114 (54%), Gaps = 9/114 (7%)
Query 1 GALDRARALYERLLDKTQHVNAFKGYATFEWKKAE---------QKDRARQVLNRGLDVC 51
G + AR LYERLL++TQHV + +A FE + AR++ R ++
Sbjct 524 GETELARQLYERLLERTQHVKVWMSFAKFEMGLSHGDSGPDAELNVQLARRIYERANEML 583
Query 52 KANGWDEDRAALLEHWLALEREGGDAQAIQRVFRLLPKKIKKRKTQKNANGVEE 105
+ G E R LLE W ER+ D+Q +Q+V +P++IKKR+ + NGVEE
Sbjct 584 RQLGDKESRVLLLEAWRDFERDASDSQEMQKVMDKMPRRIKKRQKIVSDNGVEE 637
> Hs11072091
Length=836
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 37/104 (35%), Positives = 58/104 (55%), Gaps = 8/104 (7%)
Query 4 DRARALYERLLDKTQHVNAFKGYATFEWKKAEQKD--RARQVL---NRGLDVCKANGWDE 58
+R R LY RLL +TQHV + +A FE ++ + RQ+ N+ + C+ E
Sbjct 681 ERTRNLYRRLLQRTQHVKVWISFAQFELSSGKEGSLTKCRQIYEEANKTMRNCEEK---E 737
Query 59 DRAALLEHWLALEREGGDAQAIQRVFRLLPKKIKKRKTQKNANG 102
+R LLE W + E E G A +RV +L+P+K+KKR+ + +G
Sbjct 738 ERLMLLESWRSFEEEFGTASDKERVDKLMPEKVKKRRKVQTDDG 781
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 27/56 (48%), Gaps = 1/56 (1%)
Query 3 LDRARALYERLLDKTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLDVCKANGWDE 58
+DRAR +YER + V + YA FE K A AR+V R ++ DE
Sbjct 347 VDRARTIYERFVLVHPDVKNWIKYARFEEKHA-YFAHARKVYERAVEFFGDEHMDE 401
> CE28622
Length=744
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 57/101 (56%), Gaps = 2/101 (1%)
Query 4 DRARALYERLLDKTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLDVCKANGWDEDRAAL 63
++AR LYE LL +T H+ + A FE + + AR+ R + N E+R L
Sbjct 539 EKARDLYETLLQRTNHIKVWISMAEFE-QTIGNFEGARKAFERA-NQSLENAEKEERLML 596
Query 64 LEHWLALEREGGDAQAIQRVFRLLPKKIKKRKTQKNANGVE 104
LE W E + GD +A++RV ++P+++KKR+ + +GV+
Sbjct 597 LEAWKECETKSGDQEALKRVETMMPRRVKKRRQIQTEDGVD 637
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 30/52 (57%), Gaps = 2/52 (3%)
Query 1 GALDRARALYERLLD-KTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLDVC 51
G + RAR+++ER LD + ++ + YA E + +Q + AR V +R + +
Sbjct 100 GEIQRARSVFERALDVDHRSISIWLQYAEME-MRCKQINHARNVFDRAITIM 150
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 33/133 (24%), Positives = 53/133 (39%), Gaps = 49/133 (36%)
Query 3 LDRARALYERLLD-------------KTQHVNAFKG------------------------ 25
+DRAR++Y+R L K + N + G
Sbjct 203 IDRARSVYQRFLHVHGINVQNWIKYAKFEERNGYIGNARAAYEKAMEYFGEEDINETVLV 262
Query 26 -YATFEWKKAEQKDRARQVLNRGLDVCKANGWDEDRAALLEHWLALEREGGDAQAIQRVF 84
+A FE ++ E +RAR + GLD +N +E + +H+ E++ G+ I+ V
Sbjct 263 AFALFEERQKEH-ERARGIFKYGLDNLPSNRTEE----IFKHYTQHEKKFGERVGIEDVI 317
Query 85 RLLPKKIKKRKTQ 97
I KRKTQ
Sbjct 318 ------ISKRKTQ 324
> At3g51110
Length=599
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 45/117 (38%), Positives = 62/117 (52%), Gaps = 15/117 (12%)
Query 4 DRARALYE------RLLDKTQHVNAFKGYATFEWKKAE-QKD---RARQVLNRGLDVCKA 53
+RARA++E RLLD+T+H + +A FE A+ Q+D AR V +R K
Sbjct 454 ERARAIFELAISQPRLLDRTKHYKVWLSFAKFEASAAQGQEDTIEHARAVFDRANTYYKE 513
Query 54 NGWD--EDRAALLEHWLALEREGGDAQAIQRVFRLLPKKIKKRKTQKNANGVEEVTE 108
+ + E+RA LLE WL +E GD A+Q LPKK+KKR +G E E
Sbjct 514 SKPELKEERANLLEDWLNMEASFGDVSAVQSK---LPKKLKKRMPITREDGETEYEE 567
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Query 3 LDRARALYERLLDKTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLDVCK 52
++R+R++YER + ++F YA FE K + Q AR V R +++ K
Sbjct 183 IERSRSIYERFVLCHPKASSFIRYAKFEMKNS-QVSLARIVYERAIEMLK 231
> At5g45990
Length=673
Score = 54.3 bits (129), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 48/90 (53%), Gaps = 9/90 (10%)
Query 1 GALDRARALYERLLDKTQHVNAFKGYATFEWKKAEQKD----RARQVLNRGLDVCKANG- 55
G ++ RALYERLLD+T+H + +A FE +E K+ AR + +R K
Sbjct 527 GEFEKTRALYERLLDRTKHCKVWISFAKFEASASEHKEDGIKSARVIFDRANTYYKDTTP 586
Query 56 -WDEDRAALLEHWLALER---EGGDAQAIQ 81
+E+RA LLE WL +E E GD +Q
Sbjct 587 ELEEERATLLEDWLNMETGFGELGDVSVVQ 616
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 46/91 (50%), Gaps = 3/91 (3%)
Query 3 LDRARALYERLLDKTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLDVCKANGWDEDRAA 62
++RAR++YER + V+AF YA FE K+ Q AR+V R +D AN DE+
Sbjct 195 IERARSIYERFVLCHPKVSAFIRYAKFEMKRGGQVKLAREVYERAVDKL-AN--DEEAEI 251
Query 63 LLEHWLALEREGGDAQAIQRVFRLLPKKIKK 93
L + E + + + +++ I+K
Sbjct 252 LFVSFAEFEERCKEVERARFIYKFALDHIRK 282
Score = 31.2 bits (69), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 27/48 (56%), Gaps = 2/48 (4%)
Query 3 LDRARALYERLLD-KTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLD 49
+DR R LYER L+ ++ A++ YA FE AE +RAR + +
Sbjct 459 IDRCRKLYERFLEWSPENCYAWRNYAEFEISLAET-ERARAIFELAIS 505
> At3g13210
Length=657
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 52/102 (50%), Gaps = 21/102 (20%)
Query 1 GALDRARALYERLLDKTQHVNAFKGYATFE----------------WKKAEQKDRARQVL 44
G L+R RALYERLLD+T+H + +A FE +K + RAR++
Sbjct 497 GELERTRALYERLLDRTKHCKVWVDFAKFEASAAEHKEDEEEEDAIERKKDGIKRAREIF 556
Query 45 NRGLDVCKANGWD--EDRAALLEHWLALER---EGGDAQAIQ 81
+R K + + E+RA LLE WL +E + GD + +Q
Sbjct 557 DRANTYNKDSTPELKEERAMLLEDWLNMETGFGKLGDVRVVQ 598
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Query 3 LDRARALYERLLDKTQ-HVNAFKGYATFEWKKAEQKDRARQVLNRGL 48
+DR R LYER L+ + + A++ YA FE AE +R R + +
Sbjct 429 IDRCRKLYERYLEWSPGNCYAWRKYAEFEMSLAET-ERTRAIFELAI 474
> SPBC31F10.11c
Length=674
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 46/106 (43%), Gaps = 15/106 (14%)
Query 5 RARALYERLLDKTQHVNAFKGYATFEWKKAEQKD---------------RARQVLNRGLD 49
+AR++Y++LL HV + +A FE E D RAR V L
Sbjct 522 KARSIYQQLLRTAPHVKVWISFANFEIAHLEDDDEEPPNEEVASPTAVVRARNVFENALA 581
Query 50 VCKANGWDEDRAALLEHWLALEREGGDAQAIQRVFRLLPKKIKKRK 95
+ G E+R LLE W E G + V L+P+ +KKR+
Sbjct 582 HLRQQGLKEERVVLLEAWKQFEAMHGTEDTRKHVSSLMPQVVKKRR 627
> At5g41770
Length=665
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 48/99 (48%), Gaps = 23/99 (23%)
Query 1 GALDRARALYERLLDKTQHVNAFKGYATFEWKKA------------------EQKD---R 39
G L+R RALYERLLD+T+H + +A FE A +KD R
Sbjct 500 GELERTRALYERLLDRTKHYKVWVSFAKFEASAAELEEDENEDEDQEEDVIEHKKDCIKR 559
Query 40 ARQVLNRGLDVCKANGWD--EDRAALLEHWLALEREGGD 76
AR + +R K + + E+RA LLE WL +E G+
Sbjct 560 ARAIFDRANTYYKDSTPELKEERATLLEDWLNMESSFGN 598
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 3 LDRARALYERLLDKTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLD 49
++RAR +YER + V+A+ YA FE K E R R V R +
Sbjct 195 IERARTIYERFVLCHPKVSAYIRYAKFEMKGGEVA-RCRSVYERATE 240
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 1 GALDRARALYERLLD-KTQHVNAFKGYATFEWKKAEQKDRARQVLNRGL 48
G +DR R LYER L+ ++ A+ YA E + + +RAR + +
Sbjct 430 GNMDRCRKLYERYLEWSPENCYAWSKYAELE-RSLVETERARAIFELAI 477
> At3g17040
Length=652
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 37/67 (55%), Gaps = 4/67 (5%)
Query 1 GALDRARALYE--RLLDKTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLDVCKANGWDE 58
G + RAR L++ + DK +HV A+ G+A E K+ +AR +L +GL C N +
Sbjct 251 GNVRRARELFDAATVADK-KHVAAWHGWANLEIKQG-NISKARNLLAKGLKFCGRNEYIY 308
Query 59 DRAALLE 65
ALLE
Sbjct 309 QTLALLE 315
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 39/81 (48%), Gaps = 5/81 (6%)
Query 6 ARALYERLLD-KTQHVNAFKGYATFEWKKAEQKDRARQVLNRGLDVCKANGWDEDRAALL 64
ARAL R + +H + + EWK+ AR++ R L + AN E + L
Sbjct 426 ARALLRRASELDPRHQPVWIAWGWMEWKEGNTTT-ARELYQRALSI-DAN--TESASRCL 481
Query 65 EHWLALEREGGDAQAIQRVFR 85
+ W LE+ G+ A +R+FR
Sbjct 482 QAWGVLEQRAGNLSAARRLFR 502
> CE05594
Length=1126
Score = 31.6 bits (70), Expect = 0.36, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 71 EREGGDAQAIQRVFRLLPKKIKKRKT 96
E++ G ++A++R +LLP+ I++RKT
Sbjct 1056 EKDNGHSRALRRAIQLLPRGIRERKT 1081
> 7295867
Length=1041
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 9/27 (33%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 11 ERLLDKTQHVNAFKGYATFEWKKAEQK 37
E+ LD Q + GY +F+W+ A+++
Sbjct 598 EKQLDSLQSIGESSGYESFKWRPADEE 624
Lambda K H
0.317 0.132 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160781780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40