bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2630_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
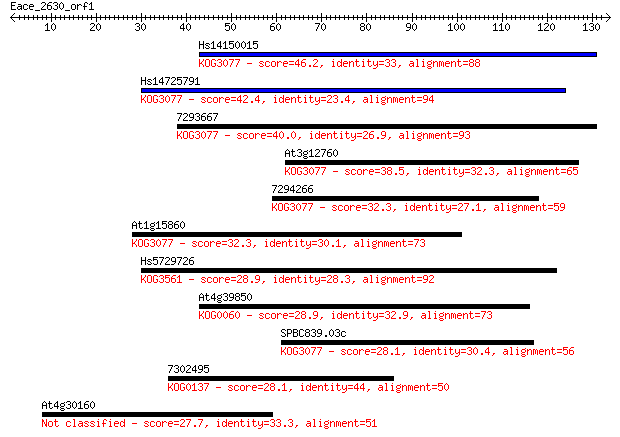
Hs14150015 46.2 1e-05
Hs14725791 42.4 2e-04
7293667 40.0 0.001
At3g12760 38.5 0.004
7294266 32.3 0.25
At1g15860 32.3 0.27
Hs5729726 28.9 2.5
At4g39850 28.9 2.9
SPBC839.03c 28.1 4.6
7302495 28.1 5.2
At4g30160 27.7 5.6
> Hs14150015
Length=237
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 48/93 (51%), Gaps = 9/93 (9%)
Query 43 AFFKVYAVEEEGGDASIKDEGLERFAADLEVGLDDIFFLLFAFYCNCRTQGIITREDFMR 102
A+F YA G D + EG+E+F D+ V ++I L+ A+ + G T+E++++
Sbjct 53 AWFYEYA----GPDEVVGPEGMEKFCEDIGVEPENIIMLVLAWKLEAESMGFFTKEEWLK 108
Query 103 GLNSLGVYTLEGLKSVVNPAREDL-----FKNI 130
G+ SL E L++ + R L FKNI
Sbjct 109 GMTSLQCDCTEKLQNKFDFLRSQLNDISSFKNI 141
> Hs14725791
Length=292
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/94 (23%), Positives = 51/94 (54%), Gaps = 0/94 (0%)
Query 30 SRKLQPKEQRLAAAFFKVYAVEEEGGDASIKDEGLERFAADLEVGLDDIFFLLFAFYCNC 89
S +++ +E+ ++ + E G D + EG+E+F D+ V +++ L+ A+ +
Sbjct 91 STRIKTEEEAFSSKRCLEWFYEYAGTDDVVGPEGMEKFCEDIGVEPENVVMLVLAWKLDA 150
Query 90 RTQGIITREDFMRGLNSLGVYTLEGLKSVVNPAR 123
+ G T +++++G+ SL T E L++ ++ R
Sbjct 151 QNMGYFTLQEWLKGMTSLQCDTTEKLRNTLDYLR 184
> 7293667
Length=233
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 51/100 (51%), Gaps = 11/100 (11%)
Query 38 QRLAAAFFKVYAVEEEGGDASIKDEGLERFAADLEVGLDDIFFLLFAFYCNCRTQGIITR 97
Q+ A+F+ Y +E ++ +G+E+F D+ V ++I L+ A+ G ++
Sbjct 34 QKRCMAWFQEYTTPDEP--ETLGPDGMEKFCEDIGVEPENIVMLVLAYKMGATQMGFFSQ 91
Query 98 EDFMRGLNSLG-------VYTLEGLKSVVNPAREDLFKNI 130
+++++GL L V L+ L+S++N FK+I
Sbjct 92 QEWLKGLTELDCDSAAKMVVKLDYLRSILNDPNS--FKSI 129
> At3g12760
Length=250
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 62 EGLERFAADLEVGLDDIFFLLFAFYCNCRTQGIITREDFMRGLNSLGVYTLEGLKSVVNP 121
EG+ DLEV DI L+ +++ N T +R++F+ GL +LGV ++ L+ +
Sbjct 78 EGISVLCNDLEVEPQDIVTLVLSWHMNAATACEFSRQEFISGLQALGVDSIGKLQEKLPF 137
Query 122 AREDL 126
R +L
Sbjct 138 MRSEL 142
> 7294266
Length=291
Score = 32.3 bits (72), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 59 IKDEGLERFAADLEVGLDDIFFLLFAFYCNCRTQGIITREDFMRGLNSLGVYTLEGLKS 117
I +G+ F DL++ D L+ A+ + Q +R++F+ G+ LG+ +++ LK+
Sbjct 80 IGSQGVIHFLEDLDLKPDSKLVLIIAWKFHAEVQCEFSRDEFINGMCDLGIDSIDKLKT 138
> At1g15860
Length=121
Score = 32.3 bits (72), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 28/73 (38%), Gaps = 3/73 (4%)
Query 28 CGSRKLQPKEQRLAAAFFKVYAVEEEGGDASIKDEGLERFAADLEVGLDDIFFLLFAFYC 87
S K KE F YA + I EG+E ++LEV DI L+ A+
Sbjct 24 SASSKASNKEMDRIDHLFNQYA---NKSSSLIDPEGIEELCSNLEVSHTDIRILMLAWKM 80
Query 88 NCRTQGIITREDF 100
QG T F
Sbjct 81 KAEKQGYFTHISF 93
> Hs5729726
Length=1412
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 26/101 (25%), Positives = 48/101 (47%), Gaps = 10/101 (9%)
Query 30 SRKLQPK----EQRLAAAFFKVYAVEEEGGDASIKDEGLERFAADLEVGLDDIFFLLFA- 84
S++LQP+ + +++ + D IK E E + DL+ LD I L +
Sbjct 762 SKELQPQVEGVDNKMSQCTSSTIPSSSQEKDPKIKTETSEEGSGDLD-NLDAILGDLTSS 820
Query 85 -FYCNCRTQG---IITREDFMRGLNSLGVYTLEGLKSVVNP 121
FY N + + T++ +G NSLG+ + + ++S+ P
Sbjct 821 DFYNNSISSNGSHLGTKQQVFQGTNSLGLKSSQSVQSIRPP 861
> At4g39850
Length=1383
Score = 28.9 bits (63), Expect = 2.9, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 33/74 (44%), Gaps = 1/74 (1%)
Query 43 AFFKVYAVEEEGGDASIK-DEGLERFAADLEVGLDDIFFLLFAFYCNCRTQGIITREDFM 101
AF+KV+ + DA + LE+ ADL L + R G+I+ FM
Sbjct 894 AFYKVFHMSGNSIDADQRLTRDLEKLTADLSGLLTGMVKPSVDILWYVRYVGLISNICFM 953
Query 102 RGLNSLGVYTLEGL 115
RG+ L Y L GL
Sbjct 954 RGVAILYTYMLLGL 967
> SPBC839.03c
Length=251
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 61 DEGLERFAADLEVGLDDIFFLLFAFYCNCRTQGIITREDFMRGLNSLGVYTLEGLK 116
D ++ F A L++ L+D LL +++ G RE F+ G +L +L+ LK
Sbjct 73 DGSVQLFTA-LDISLEDPETLLVSYFLKSPRMGEFHRESFVEGALNLSTTSLDQLK 127
> 7302495
Length=655
Score = 28.1 bits (61), Expect = 5.2, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 27/66 (40%), Gaps = 16/66 (24%)
Query 36 KEQRLAAAFFKVYAVEEE-----------GGDASIKDEGLERFAADLEV-----GLDDIF 79
K+ L AA K+YA E GG + D GLER DL + G +DI
Sbjct 410 KDYHLEAAISKIYASESAWYVCDEAIQILGGMGYMVDNGLERVLRDLRIFRIFEGTNDIL 469
Query 80 FLLFAF 85
L A
Sbjct 470 RLFIAL 475
> At4g30160
Length=974
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 8 DAVAAAAAAAATDAAGAGGVCGSRKLQPKEQRLAAAFFKVYAVEEEGGDAS 58
+A AA DAA G R++Q E ++FK + +EGG AS
Sbjct 77 EAGTAAVKTVELDAALGGRAVQYREVQGHETEKFLSYFKPCIIPQEGGVAS 127
Lambda K H
0.321 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1356426142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40