bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2637_orf1
Length=128
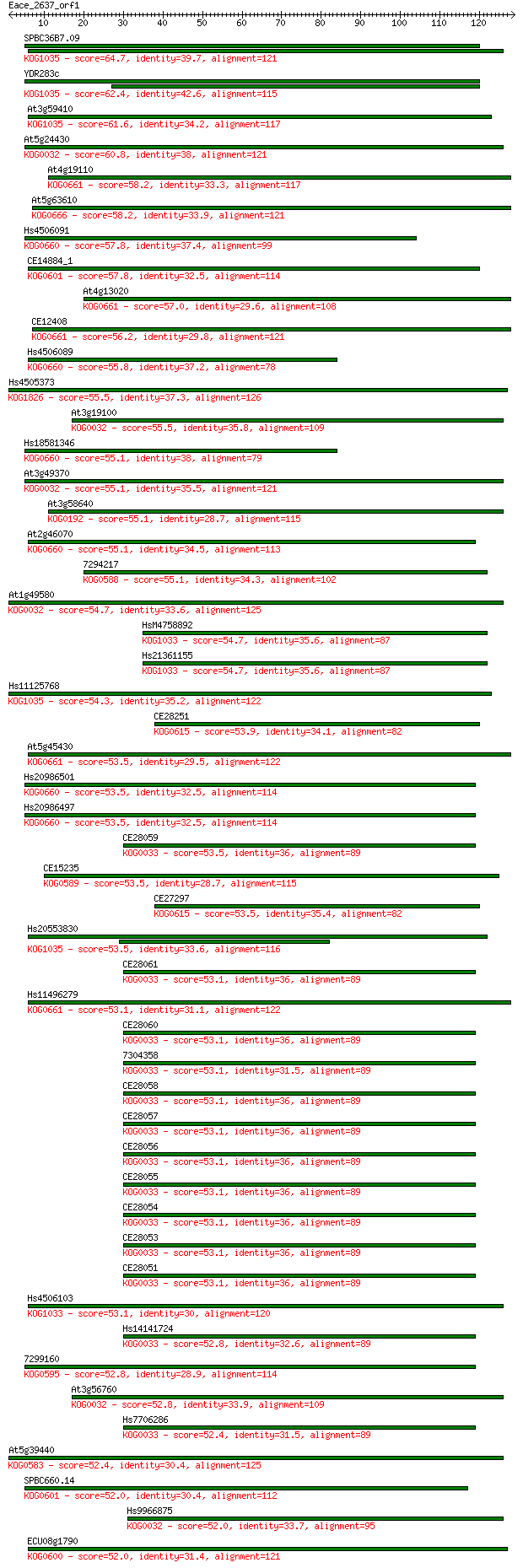
Score E
Sequences producing significant alignments: (Bits) Value
SPBC36B7.09 64.7 4e-11
YDR283c 62.4 2e-10
At3g59410 61.6 3e-10
At5g24430 60.8 7e-10
At4g19110 58.2 3e-09
At5g63610 58.2 4e-09
Hs4506091 57.8 4e-09
CE14884_1 57.8 5e-09
At4g13020 57.0 8e-09
CE12408 56.2 2e-08
Hs4506089 55.8 2e-08
Hs4505373 55.5 2e-08
At3g19100 55.5 3e-08
Hs18581346 55.1 3e-08
At3g49370 55.1 3e-08
At3g58640 55.1 4e-08
At2g46070 55.1 4e-08
7294217 55.1 4e-08
At1g49580 54.7 4e-08
HsM4758892 54.7 5e-08
Hs21361155 54.7 5e-08
Hs11125768 54.3 5e-08
CE28251 53.9 8e-08
At5g45430 53.5 9e-08
Hs20986501 53.5 9e-08
Hs20986497 53.5 1e-07
CE28059 53.5 1e-07
CE15235 53.5 1e-07
CE27297 53.5 1e-07
Hs20553830 53.5 1e-07
CE28061 53.1 1e-07
Hs11496279 53.1 1e-07
CE28060 53.1 1e-07
7304358 53.1 1e-07
CE28058 53.1 1e-07
CE28057 53.1 1e-07
CE28056 53.1 1e-07
CE28055 53.1 1e-07
CE28054 53.1 1e-07
CE28053 53.1 1e-07
CE28051 53.1 1e-07
Hs4506103 53.1 1e-07
Hs14141724 52.8 1e-07
7299160 52.8 1e-07
At3g56760 52.8 2e-07
Hs7706286 52.4 2e-07
At5g39440 52.4 2e-07
SPBC660.14 52.0 3e-07
Hs9966875 52.0 3e-07
ECU08g1790 52.0 3e-07
> SPBC36B7.09
Length=903
Score = 64.7 bits (156), Expect = 4e-11, Method: Composition-based stats.
Identities = 46/137 (33%), Positives = 70/137 (51%), Gaps = 27/137 (19%)
Query 5 SVFVQMEYYE-LALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFL 63
++++QMEY E L+LQ + ++ + + ++ RLF Q+LEA+ +H G HR L P N+FL
Sbjct 723 TLYIQMEYCEKLSLQDIIRD--KIPVDEMWRLFRQILEALAYIHSRGMMHRDLKPGNIFL 780
Query 64 VEGSEGLIMKVGDFGL----------------RQLV--PLPPDSPFASFVAPEV---HKG 102
E +K+GDFGL RQ L A +VAPE+ G
Sbjct 781 DENRN---VKLGDFGLATENENYQDNNDKWKNRQSADEDLTTGVGTALYVAPELLSRRNG 837
Query 103 HPFTDKADMYSIGMLFL 119
+ K DMYS+G++
Sbjct 838 VRYDAKVDMYSLGIILF 854
Score = 43.5 bits (101), Expect = 1e-04, Method: Composition-based stats.
Identities = 38/136 (27%), Positives = 56/136 (41%), Gaps = 18/136 (13%)
Query 6 VFVQMEYY-ELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLV 64
++V EY + L L + TL + VR +LE + +HR G H++LH NV L
Sbjct 324 LYVLQEYSPKFTLFSLLQTVLTLDVETVRAFSNNILEGLAELHRLGISHKSLHLDNVVLF 383
Query 65 EGSEGLIMKVGDFGL-RQLVPLPPDSPF------------ASFVAPEVHKGH--PFTDKA 109
K+ DFG R L + PF PEV + + K
Sbjct 384 HSGHRTFAKLMDFGFTRTLRDMNASHPFNINSQSITNILPEGLYPPEVSESSFAAASRKT 443
Query 110 DMYSIGMLFLGLWLLC 125
D++ G+L L +LC
Sbjct 444 DIWCFGLLVLQ--MLC 457
> YDR283c
Length=1659
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 49/138 (35%), Positives = 69/138 (50%), Gaps = 27/138 (19%)
Query 5 SVFVQMEYYE--LALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVF 62
++F+QMEY E + E + + RLF Q+LEA+ +H G HR L P N+F
Sbjct 783 TLFIQMEYCENRTLYDLIHSENLNQQRDEYWRLFRQILEALSYIHSQGIIHRDLKPMNIF 842
Query 63 LVEGSEGLIMKVGDFGL-----RQLVPLPPDS---PFAS-----------FVAPEV--HK 101
+ E +K+GDFGL R L L DS P +S +VA EV
Sbjct 843 IDESRN---VKIGDFGLAKNVHRSLDILKLDSQNLPGSSDNLTSAIGTAMYVATEVLDGT 899
Query 102 GHPFTDKADMYSIGMLFL 119
GH + +K DMYS+G++F
Sbjct 900 GH-YNEKIDMYSLGIIFF 916
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 23/109 (21%), Positives = 54/109 (49%), Gaps = 17/109 (15%)
Query 27 LKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSE------GLIMKVGDFGLR 80
+ +A R ++LLE + +H+ G H+ ++ V LV+ ++ L+ + +
Sbjct 365 VNLATARIWMIRLLEGLEAIHKLGIVHKCINLETVILVKDADFGSTIPKLVHSTYGYTVL 424
Query 81 QLVPLPPD--------SPFASFVAPEVHKGHPFTDK--ADMYSIGMLFL 119
++ P+ SP ++++APE+ K + + D++ +G+LF+
Sbjct 425 NMLSRYPNKNGSSVELSP-STWIAPELLKFNNAKPQRLTDIWQLGVLFI 472
> At3g59410
Length=1271
Score = 61.6 bits (148), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 40/138 (28%), Positives = 66/138 (47%), Gaps = 24/138 (17%)
Query 6 VFVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVE 65
+++QMEY L+++ + L Q++E + +H G HR P+N+F
Sbjct 566 LYIQMEYCPRTLRQVFESYNHFDKDFAWHLIRQIVEGLAHIHGQGIIHRDFTPNNIFFDA 625
Query 66 GSEGLIMKVGDFGLR---QLVPLPPDSPFAS-----------------FVAPEVHKGHPF 105
++ +K+GDFGL +L L D F++ + APE+ + P
Sbjct 626 RND---IKIGDFGLAKFLKLEQLDQDGGFSTDVAGSGVDSTGQAGTYFYTAPEIEQDWPK 682
Query 106 TD-KADMYSIGMLFLGLW 122
D KADMYS+G++F LW
Sbjct 683 IDEKADMYSLGVVFFELW 700
> At5g24430
Length=594
Score = 60.8 bits (146), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 46/128 (35%), Positives = 63/128 (49%), Gaps = 11/128 (8%)
Query 5 SVFVQMEYYELA--LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVF 62
+VFV ME E L R+ G D +R+ +Q+L A H G HR L P N
Sbjct 219 NVFVVMELCEGGELLDRILARGGRYPEVDAKRILVQILSATAFFHLQGVVHRDLKPENFL 278
Query 63 LVEGSEGLIMKVGDFGLRQLV----PLPPDSPFASFVAPEV-HKGHPFTDKADMYSIGML 117
+E I+KV DFGL + L A +VAPEV H+ ++ +ADM+SIG+
Sbjct 279 FTSRNEDAILKVIDFGLSDFIRYDQRLNDVVGSAYYVAPEVLHRS--YSTEADMWSIGV- 335
Query 118 FLGLWLLC 125
+ LLC
Sbjct 336 -ISYILLC 342
> At4g19110
Length=256
Score = 58.2 bits (139), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 67/123 (54%), Gaps = 10/123 (8%)
Query 11 EYYELALQRLTKEGRTL-KIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEG 69
EY E L +L K+ + L AD++ Q+ + + +H+ G HR L P N+ + +
Sbjct 46 EYMECNLYQLMKDRQKLFAEADIKNWCFQVFQGLSYMHQRGYFHRDLKPENLLVSKD--- 102
Query 70 LIMKVGDFGL-RQLVPLPPDSPFAS---FVAPEV-HKGHPFTDKADMYSIGMLFLGLWLL 124
I+K+ DFGL R++ PP + + S + APEV + + +T K DM+++G + L L
Sbjct 103 -IIKIADFGLAREVNSSPPFTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMAELLSL 161
Query 125 CPV 127
P+
Sbjct 162 RPI 164
> At5g63610
Length=470
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 41/130 (31%), Positives = 64/130 (49%), Gaps = 9/130 (6%)
Query 7 FVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLV-E 65
+ + + YE+ K G +L V+ L QLL + +H HR L PSN+ ++ +
Sbjct 106 YAEYDLYEIIRHHRDKVGHSLNTYTVKSLLWQLLNGLNYLHSNWIIHRDLKPSNILVMGD 165
Query 66 GSEGLIMKVGDFGLRQL--VPLPPDSPFASFV-----APEVHKGHP-FTDKADMYSIGML 117
E I+K+ DFGL ++ PL P S V APE+ G +T DM+++G +
Sbjct 166 AEEHGIVKIADFGLARIYQAPLKPLSDNGVVVTIWYRAPELLLGSKHYTSAVDMWAVGCI 225
Query 118 FLGLWLLCPV 127
F L L P+
Sbjct 226 FAELLTLKPL 235
> Hs4506091
Length=721
Score = 57.8 bits (138), Expect = 4e-09, Method: Composition-based stats.
Identities = 37/100 (37%), Positives = 56/100 (56%), Gaps = 16/100 (16%)
Query 5 SVFVQMEYYELALQRLTKEGRTLKIADVRRLFM-QLLEAIVCVHRAGACHRALHPSNVFL 63
SV++ EY E L + ++G L+ + RLFM QLL + +H A HR L P+N+F+
Sbjct 103 SVYIVQEYMETDLANVLEQGPLLE--EHARLFMYQLLRGLKYIHSANVLHRDLKPANLFI 160
Query 64 VEGSEGLIMKVGDFGLRQLVPLPPDSPFASFVAPEVHKGH 103
+E L++K+GDFGL +++ P S HKGH
Sbjct 161 --NTEDLVLKIGDFGLARIM-----DPHYS------HKGH 187
> CE14884_1
Length=415
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 37/119 (31%), Positives = 63/119 (52%), Gaps = 9/119 (7%)
Query 6 VFVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVE 65
+++Q E + +L + E L ++ +F+ LL+A+ +H H + P N+FL
Sbjct 179 LYIQTELCDQSLLKYCTEKHALPEDEIWNIFVDLLQAVHHLHSNDMIHDDIKPENIFL-- 236
Query 66 GSEGLIMKVGDFGLRQLVPLPPDSPFAS-----FVAPEVHKGHPFTDKADMYSIGMLFL 119
++ +I K+GDFGL + P D A ++APEV G P T +D++S+GM L
Sbjct 237 -TKDMICKLGDFGLVINLKNPNDVKSAEEGDSKYLAPEVLNGRP-TKSSDIFSLGMTIL 293
> At4g13020
Length=443
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 59/113 (52%), Gaps = 9/113 (7%)
Query 20 LTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGL 79
+ + R ++R Q+L+ + +H+ G HR L P N+ + I+K+ DFGL
Sbjct 98 MKERERPFSEGEIRSFMSQMLQGLAHMHKNGYFHRDLKPENLLVTNN----ILKIADFGL 153
Query 80 -RQLVPLPPDSPFAS---FVAPEV-HKGHPFTDKADMYSIGMLFLGLWLLCPV 127
R++ +PP + + S + APEV + +T DM+++G + L+ L P+
Sbjct 154 AREVASMPPYTEYVSTRWYRAPEVLLQSSLYTPAVDMWAVGAILAELYALTPL 206
> CE12408
Length=527
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 36/126 (28%), Positives = 67/126 (53%), Gaps = 12/126 (9%)
Query 7 FVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEG 66
F+Q YEL R R + +R + Q+L+ + +H+ G HR + P N+ + G
Sbjct 122 FMQENLYELMKDR----DRYFPESVIRNIIYQVLQGLAFMHKNGFFHRDMKPENI-MCNG 176
Query 67 SEGLIMKVGDFGL-RQLVPLPPDSPFAS---FVAPEV-HKGHPFTDKADMYSIGMLFLGL 121
+E ++K+ DFGL R++ PP + + S + APE+ + + DM+++G + L
Sbjct 177 TE--LVKIADFGLAREIRSKPPYTDYVSTRWYRAPEILLRSTSYNSPIDMWALGCIMAEL 234
Query 122 WLLCPV 127
++L P+
Sbjct 235 YILRPL 240
> Hs4506089
Length=557
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 6 VFVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVE 65
++ EY E L RL ++G TL + QLL + +H A HR L P+N+F+
Sbjct 101 AYIVQEYMETDLARLLEQG-TLAEEHAKLFMYQLLRGLKYIHSANVLHRDLKPANIFI-- 157
Query 66 GSEGLIMKVGDFGLRQLV 83
+E L++K+GDFGL ++V
Sbjct 158 STEDLVLKIGDFGLARIV 175
> Hs4505373
Length=445
Score = 55.5 bits (132), Expect = 2e-08, Method: Composition-based stats.
Identities = 47/143 (32%), Positives = 74/143 (51%), Gaps = 22/143 (15%)
Query 1 QTQLSVFVQMEYYE---LA--LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAG----- 50
+T ++++ MEY E LA + + TKE + L V R+ QL A+ HR
Sbjct 77 RTNTTLYIVMEYCEGGDLASVITKGTKERQYLDEEFVLRVMTQLTLALKECHRRSDGGHT 136
Query 51 ACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDSPFAS-------FVAPEVHKGH 103
HR L P+NVFL +G + + K+GDFGL ++ L D+ FA +++PE
Sbjct 137 VLHRDLKPANVFL-DGKQNV--KLGDFGLARI--LNHDTSFAKTFVGTPYYMSPEQMNRM 191
Query 104 PFTDKADMYSIGMLFLGLWLLCP 126
+ +K+D++S+G L L L P
Sbjct 192 SYNEKSDIWSLGCLLYELCALMP 214
> At3g19100
Length=599
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 55/116 (47%), Gaps = 13/116 (11%)
Query 17 LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGD 76
L R+ G D + + +Q+L + H G HR L P N E ++KV D
Sbjct 234 LDRILARGGKYSEDDAKAVLIQILNVVAFCHLQGVVHRDLKPENFLYTSKEENSMLKVID 293
Query 77 FGLRQLVPLPPDSPF------ASFVAPEV-HKGHPFTDKADMYSIGMLFLGLWLLC 125
FGL V PD A +VAPEV H+ +T +AD++SIG+ + LLC
Sbjct 294 FGLSDFV--RPDERLNDIVGSAYYVAPEVLHRS--YTTEADVWSIGV--IAYILLC 343
> Hs18581346
Length=377
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 51/80 (63%), Gaps = 5/80 (6%)
Query 5 SVFVQMEYYELALQRLTKEGRTLKIADVRRLFM-QLLEAIVCVHRAGACHRALHPSNVFL 63
SV++ EY + L ++ ++G L+ + RLFM QLL + +H A HR L P+N+F+
Sbjct 133 SVYIVQEYMKTDLSKVLEQGPLLE--EHARLFMYQLLRGLKYIHSANVLHRDLKPTNLFI 190
Query 64 VEGSEGLIMKVGDFGLRQLV 83
+E L++K+GDFGL +++
Sbjct 191 --NTEDLVLKIGDFGLARIM 208
> At3g49370
Length=594
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 62/128 (48%), Gaps = 11/128 (8%)
Query 5 SVFVQMEYYELA--LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVF 62
+VFV ME E L + G A+ +R+ +Q+L A H G HR L P N
Sbjct 218 NVFVVMELCEGGELLDSILARGGRYPEAEAKRILVQILSATAFFHLQGVVHRDLKPENFL 277
Query 63 LVEGSEGLIMKVGDFGLRQLV----PLPPDSPFASFVAPEV-HKGHPFTDKADMYSIGML 117
+E ++KV DFGL L A +VAPEV H+ ++ +AD++SIG+
Sbjct 278 FTSKNEDAVLKVIDFGLSDYARFDQRLNDVVGSAYYVAPEVLHRS--YSTEADIWSIGV- 334
Query 118 FLGLWLLC 125
+ LLC
Sbjct 335 -ISYILLC 341
> At3g58640
Length=816
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 33/126 (26%), Positives = 64/126 (50%), Gaps = 19/126 (15%)
Query 11 EYYELA----LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEG 66
EY E+ L L+ + + L ++ + ++C+HR G HR + +N L
Sbjct 635 EYMEMGSLYYLLHLSGQKKRLSWRRKLKMLRDICRGLMCIHRMGIVHRDIKSANCLL--- 691
Query 67 SEGLIMKVGDFGLRQLVP-------LPPDSPFASFVAPEVHKGHPFTDKADMYSIGMLFL 119
S +K+ DFGL +++ + +P ++APE+ + PF++K D++S+G++
Sbjct 692 SNKWTVKICDFGLSRIMTGTTMRDTVSAGTP--EWMAPELIRNEPFSEKCDIFSLGVI-- 747
Query 120 GLWLLC 125
+W LC
Sbjct 748 -MWELC 752
> At2g46070
Length=406
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 39/121 (32%), Positives = 59/121 (48%), Gaps = 14/121 (11%)
Query 6 VFVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVE 65
V++ E + LQR+ + +TL R L QLL + VH A HR L PSNV L
Sbjct 152 VYIVYELMDTDLQRILRSNQTLTSDQCRFLVYQLLRGLKYVHSANILHRDLRPSNVLLNS 211
Query 66 GSEGLIMKVGDFGLRQLVPLPPDSPFAS-------FVAPE-VHKGHPFTDKADMYSIGML 117
+E +K+GDFGL + D+ F + + APE + +T D++S+G +
Sbjct 212 KNE---LKIGDFGLARTT---SDTDFMTEYVVTRWYRAPELLLNCSEYTAAIDIWSVGCI 265
Query 118 F 118
Sbjct 266 L 266
> 7294217
Length=851
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 35/107 (32%), Positives = 53/107 (49%), Gaps = 9/107 (8%)
Query 20 LTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGL 79
L K+GR L + R+ F Q++ A+ H CHR L P N+ L E + +K+ DFG+
Sbjct 106 LVKKGR-LTPKEARKFFRQIISALDFCHSHSICHRDLKPENLLLDEKNN---IKIADFGM 161
Query 80 RQLVP----LPPDSPFASFVAPEVHKGHPFTD-KADMYSIGMLFLGL 121
L P L + PEV +G + KAD++S G++ L
Sbjct 162 ASLQPAGSMLETSCGSPHYACPEVIRGEKYDGRKADVWSCGVILYAL 208
> At1g49580
Length=606
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 42/134 (31%), Positives = 61/134 (45%), Gaps = 15/134 (11%)
Query 1 QTQLSVFVQMEYYELA--LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHP 58
+ +V++ ME E L R+ G D + + +Q+L + H G HR L P
Sbjct 222 EDNANVYIAMELCEGGELLDRILARGGKYSENDAKPVIIQILNVVAFCHFQGVVHRDLKP 281
Query 59 SNVFLVEGSEGLIMKVGDFGLRQLVPLPPDSPF------ASFVAPEV-HKGHPFTDKADM 111
N E +K DFGL V PD A +VAPEV H+ +T +AD+
Sbjct 282 ENFLYTSKEENSQLKAIDFGLSDFV--RPDERLNDIVGSAYYVAPEVLHRS--YTTEADV 337
Query 112 YSIGMLFLGLWLLC 125
+SIG+ + LLC
Sbjct 338 WSIGV--IAYILLC 349
> HsM4758892
Length=1115
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 31/104 (29%), Positives = 52/104 (50%), Gaps = 20/104 (19%)
Query 35 LFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQ-----------LV 83
+F+Q+ EA+ +H G HR L PSN+F ++KVGDFGL L
Sbjct 916 IFLQIAEAVEFLHSKGLMHRDLKPSNIFFTMDD---VVKVGDFGLVTAMDQDEEEQTVLT 972
Query 84 PLPPDSPFAS------FVAPEVHKGHPFTDKADMYSIGMLFLGL 121
P+P + +++PE G+ ++ K D++S+G++ L
Sbjct 973 PMPAYARHTGQVGTKLYMSPEQIHGNSYSHKVDIFSLGLILFEL 1016
> Hs21361155
Length=1115
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 31/104 (29%), Positives = 52/104 (50%), Gaps = 20/104 (19%)
Query 35 LFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQ-----------LV 83
+F+Q+ EA+ +H G HR L PSN+F ++KVGDFGL L
Sbjct 916 IFLQIAEAVEFLHSKGLMHRDLKPSNIFFTMDD---VVKVGDFGLVTAMDQDEEEQTVLT 972
Query 84 PLPPDSPFAS------FVAPEVHKGHPFTDKADMYSIGMLFLGL 121
P+P + +++PE G+ ++ K D++S+G++ L
Sbjct 973 PMPAYARHTGQVGTKLYMSPEQIHGNSYSHKVDIFSLGLILFEL 1016
> Hs11125768
Length=630
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 43/152 (28%), Positives = 72/152 (47%), Gaps = 32/152 (21%)
Query 1 QTQLSVFVQMEYYELALQ----RLTKEGRTLK--------IADV-RRLFMQLLEAIVCVH 47
Q L + +QM+ EL+L K GR +A+V ++F +L+E + +H
Sbjct 375 QYHLMLHIQMQLCELSLWDWIVERNKRGREYVDESACPYVMANVATKIFQELVEGVFYIH 434
Query 48 RAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDSPFAS-------------- 93
G HR L P N+FL G + + K+GDFGL L ++ + +
Sbjct 435 NMGIVHRDLKPRNIFL-HGPDQQV-KIGDFGLACTDILQKNTDWTNRNGKRTPTHTSRVG 492
Query 94 ---FVAPEVHKGHPFTDKADMYSIGMLFLGLW 122
+ +PE +G + K+DMYS+G++ L L+
Sbjct 493 TCLYASPEQLEGSEYDAKSDMYSLGVVLLELF 524
> CE28251
Length=469
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 51/87 (58%), Gaps = 5/87 (5%)
Query 38 QLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGL--RQLVPLPPDSPFASFV 95
QL++A++ +H G CHR + P N+ +E I+K+ DFG+ + + S+
Sbjct 283 QLIDAVLYLHSVGICHRDIKPENILCSNKAERCILKLTDFGMAKNSVNRMKTHCGTPSYC 342
Query 96 APEV--HKGHPFTDKADMYSIG-MLFL 119
APE+ ++G +T K D++S+G +LF+
Sbjct 343 APEIVANQGVEYTPKVDIWSLGCVLFI 369
> At5g45430
Length=530
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 36/128 (28%), Positives = 67/128 (52%), Gaps = 10/128 (7%)
Query 6 VFVQMEYYELALQRLTKE-GRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLV 64
++ EY E L +L K+ + +D+R Q+ + + +H+ G HR L P N+ +
Sbjct 83 LYFVFEYMECNLYQLMKDRPKHFAESDIRNWCFQVFQGLSYMHQRGYFHRDLKPENLLVS 142
Query 65 EGSEGLIMKVGDFGL-RQLVPLPPDSPFAS---FVAPEV-HKGHPFTDKADMYSIGMLFL 119
+ ++K+ D GL R++ PP + + S + APEV + + +T K DM+++G +
Sbjct 143 KD----VIKIADLGLAREIDSSPPYTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGAIMA 198
Query 120 GLWLLCPV 127
L L P+
Sbjct 199 ELLSLRPL 206
> Hs20986501
Length=816
Score = 53.5 bits (127), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 61/124 (49%), Gaps = 13/124 (10%)
Query 5 SVFVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLV 64
SV+V ++ E L ++ + L + VR QLL + +H A HR L PSN+ +
Sbjct 132 SVYVVLDLMESDLHQIIHSSQPLTLEHVRYFLYQLLRGLKYMHSAQVIHRDLKPSNLLVN 191
Query 65 EGSEGLIMKVGDFGL-RQLVPLPPDSPF--ASFVAPEVHKG-------HPFTDKADMYSI 114
E E +K+GDFG+ R L P + + +VA ++ H +T D++S+
Sbjct 192 ENCE---LKIGDFGMARGLCTSPAEHQYFMTEYVATRWYRAPELMLSLHEYTQAIDLWSV 248
Query 115 GMLF 118
G +F
Sbjct 249 GCIF 252
> Hs20986497
Length=816
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 61/124 (49%), Gaps = 13/124 (10%)
Query 5 SVFVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLV 64
SV+V ++ E L ++ + L + VR QLL + +H A HR L PSN+ +
Sbjct 132 SVYVVLDLMESDLHQIIHSSQPLTLEHVRYFLYQLLRGLKYMHSAQVIHRDLKPSNLLVN 191
Query 65 EGSEGLIMKVGDFGL-RQLVPLPPDSPF--ASFVAPEVHKG-------HPFTDKADMYSI 114
E E +K+GDFG+ R L P + + +VA ++ H +T D++S+
Sbjct 192 ENCE---LKIGDFGMARGLCTSPAEHQYFMTEYVATRWYRAPELMLSLHEYTQAIDLWSV 248
Query 115 GMLF 118
G +F
Sbjct 249 GCIF 252
> CE28059
Length=350
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> CE15235
Length=357
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/129 (25%), Positives = 64/129 (49%), Gaps = 16/129 (12%)
Query 10 MEYYEL-ALQRLTKEGRTLKIADVRRLF---------MQLLEAIVCVHRAGACHRALHPS 59
M+Y E L+RL + R +K +++R F Q+L A+ +H+ HR L P
Sbjct 82 MQYAEGGTLERLINDQRAIKDSNMREYFPEKTVLDYFTQILIALNHMHQKNIVHRDLKPQ 141
Query 60 NVFLVEGSEGLIMKVGDFGLRQLVPLPPDSPFA----SFVAPEVHKGHPFTDKADMYSIG 115
N+ + ++K+ DFG+ + + + ++++PE+ + P+ K+DM+S+G
Sbjct 142 NILM--NRRKTVLKLSDFGISKELGTKSAASTVIGTPNYLSPEICESRPYNQKSDMWSLG 199
Query 116 MLFLGLWLL 124
+ L L
Sbjct 200 CVLYELLQL 208
> CE27297
Length=476
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 53/87 (60%), Gaps = 5/87 (5%)
Query 38 QLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQ--LVPLPPDSPFASFV 95
QL++AI+ +H G CHR + P N+ + +E I+K+ DFG+ + + + S+
Sbjct 284 QLIDAILYLHSVGICHRDIKPENILCSDKAERCILKLTDFGMAKNSVNRMKTRCGTPSYN 343
Query 96 APEV--HKGHPFTDKADMYSIG-MLFL 119
APE+ ++G +T K D++S+G +LF+
Sbjct 344 APEIVANEGVEYTPKVDIWSLGCVLFI 370
> Hs20553830
Length=1528
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/142 (27%), Positives = 63/142 (44%), Gaps = 29/142 (20%)
Query 6 VFVQMEYYELALQRLT-KEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLV 64
+++QMEY E + R T +G + RLF ++L+ + +H G HR L P N+FL
Sbjct 677 LYIQMEYCEKSTLRDTIDQGLYRDTVRLWRLFREILDGLAYIHEKGMIHRDLKPVNIFLD 736
Query 65 EGSEGLIMKVGDFGLR-----------------QLVPLPPDSPF------ASFVAPEVHK 101
+K+GDFGL L+ P A +V+PEV
Sbjct 737 SDDH---VKIGDFGLATDHLAFSADSKQDDQTGDLIKSDPSGHLTGMVGTALYVSPEVQG 793
Query 102 G--HPFTDKADMYSIGMLFLGL 121
+ K D++S+G++F +
Sbjct 794 STKSAYNQKVDLFSLGIIFFEM 815
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 27/53 (50%), Gaps = 3/53 (5%)
Query 29 IADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQ 81
+ +RR QLL + +H H+ L SNV + +EG + K+ D+ + +
Sbjct 278 VHQLRRYTAQLLSGLDYLHSNSVVHKVLSASNVLV--DAEGTV-KITDYSISK 327
> CE28061
Length=302
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> Hs11496279
Length=623
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 38/128 (29%), Positives = 69/128 (53%), Gaps = 9/128 (7%)
Query 6 VFVQMEYYELALQRLTKEGRTLKIADV-RRLFMQLLEAIVCVHRAGACHRALHPSNVFLV 64
++ EY + L +L K+ L V R + Q+L+ + +H+ G HR + P N+ L
Sbjct 75 LYFIFEYMKENLYQLMKDRNKLFPESVIRNIMYQILQGLAFIHKHGFFHRDMKPENL-LC 133
Query 65 EGSEGLIMKVGDFGL-RQLVPLPPDSPFAS---FVAPEV-HKGHPFTDKADMYSIGMLFL 119
G E ++K+ DFGL R+L PP + + S + APEV + ++ D++++G +
Sbjct 134 MGPE--LVKIADFGLARELRSQPPYTDYVSTRWYRAPEVLLRSSVYSSPIDVWAVGSIMA 191
Query 120 GLWLLCPV 127
L++L P+
Sbjct 192 ELYMLRPL 199
> CE28060
Length=302
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> 7304358
Length=493
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 5/94 (5%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE++ H+ G HR L P N+ L ++G +K+ DFGL V +
Sbjct 111 ADASHCIQQILESVNHCHQNGVVHRDLKPENLLLASKAKGAAVKLADFGLAIEVQGDHQA 170
Query 90 PFA-----SFVAPEVHKGHPFTDKADMYSIGMLF 118
F +++PEV K P+ D+++ G++
Sbjct 171 WFGFAGTPGYLSPEVLKKEPYGKSVDIWACGVIL 204
> CE28058
Length=571
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> CE28057
Length=520
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> CE28056
Length=518
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> CE28055
Length=533
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> CE28054
Length=482
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> CE28053
Length=720
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> CE28051
Length=559
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE+I H G HR L P N+ L ++G +K+ DFGL V DS
Sbjct 109 ADASHCIQQILESIAYCHSNGIVHRDLKPENLLLASKAKGAAVKLADFGLAIEV---NDS 165
Query 90 P----FA---SFVAPEVHKGHPFTDKADMYSIGMLF 118
FA +++PEV K P++ D+++ G++
Sbjct 166 EAWHGFAGTPGYLSPEVLKKDPYSKPVDIWACGVIL 201
> Hs4506103
Length=551
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 36/129 (27%), Positives = 66/129 (51%), Gaps = 14/129 (10%)
Query 6 VFVQMEYYELA-LQRLTKEGRTLKIADVR--RLFMQLLEAIVCVHRAGACHRALHPSNVF 62
+F+QME+ + L++ ++ R K+ V LF Q+ + + +H HR L PSN+F
Sbjct 362 LFIQMEFCDKGTLEQWIEKRRGEKLDKVLALELFEQITKGVDYIHSKKLIHRDLKPSNIF 421
Query 63 LVEGSEGLIMKVGDFGLRQLVPLPPDSPFA------SFVAPEVHKGHPFTDKADMYSIGM 116
LV+ + +K+GDFGL + L D +++PE + + D+Y++G+
Sbjct 422 LVDTKQ---VKIGDFGL--VTSLKNDGKRTRSKGTLRYMSPEQISSQDYGKEVDLYALGL 476
Query 117 LFLGLWLLC 125
+ L +C
Sbjct 477 ILAELLHVC 485
> Hs14141724
Length=499
Score = 52.8 bits (125), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/94 (30%), Positives = 46/94 (48%), Gaps = 5/94 (5%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LE++ H G HR L P N+ L S+G +K+ DFGL V +
Sbjct 111 ADASHCIQQILESVNHCHLNGIVHRDLKPENLLLASKSKGAAVKLADFGLAIEVQGDQQA 170
Query 90 PFA-----SFVAPEVHKGHPFTDKADMYSIGMLF 118
F +++PEV + P+ DM++ G++
Sbjct 171 WFGFAGTPGYLSPEVLRKDPYGKPVDMWACGVIL 204
> 7299160
Length=520
Score = 52.8 bits (125), Expect = 1e-07, Method: Composition-based stats.
Identities = 33/121 (27%), Positives = 57/121 (47%), Gaps = 9/121 (7%)
Query 5 SVFVQMEYYELA-LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFL 63
++++ +EY L + + L + R QL A+ + H L P N+ L
Sbjct 80 NIYIVLEYCNAGNLSAFIRTKKALPESTCRYFLRQLAAAVQYMRANDVSHFDLKPQNLLL 139
Query 64 VEGSEGLIMKVGDFGLRQLVPLP------PDSPFASFVAPEVHKGHPFTDKADMYSIGML 117
G+ + +KV DFG Q + L SP ++APE+ + H + KAD++SIG++
Sbjct 140 TRGANNVSLKVADFGFAQHLKLGEINQQLKGSPL--YMAPEIVRKHQYDAKADLWSIGVI 197
Query 118 F 118
Sbjct 198 L 198
> At3g56760
Length=577
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 54/116 (46%), Gaps = 13/116 (11%)
Query 17 LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGD 76
L ++ + G D +++ +Q+L + H G HR L P N E +K D
Sbjct 214 LDKILQRGGKYSEVDAKKVMIQILSVVAYCHLQGVVHRDLKPENFLFTTKDESSPLKAID 273
Query 77 FGLRQLVPLPPDSPF------ASFVAPEV-HKGHPFTDKADMYSIGMLFLGLWLLC 125
FGL V PD A +VAPEV H+ + +ADM+SIG+ + LLC
Sbjct 274 FGLSDYV--RPDERLNDIVGSAYYVAPEVLHR--TYGTEADMWSIGV--IAYILLC 323
> Hs7706286
Length=489
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 5/94 (5%)
Query 30 ADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQLVPLPPDS 89
AD Q+LEA++ H+ G HR L P N+ L +G +K+ DFGL V +
Sbjct 110 ADASHCIQQILEAVLHCHQMGVVHRDLKPENLLLASKLKGAAVKLADFGLAIEVEGEQQA 169
Query 90 PFA-----SFVAPEVHKGHPFTDKADMYSIGMLF 118
F +++PEV + P+ D+++ G++
Sbjct 170 WFGFAGTPGYLSPEVLRKDPYGKPVDLWACGVIL 203
> At5g39440
Length=494
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 38/131 (29%), Positives = 62/131 (47%), Gaps = 12/131 (9%)
Query 1 QTQLSVFVQMEYYELA--LQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHP 58
+T ++V MEY + + ++G+ L+ + R LF Q++ + HR HR L P
Sbjct 87 ETPNDIYVVMEYVKSGELFDYIVEKGK-LQEDEARHLFQQIISGVEYCHRNMIVHRDLKP 145
Query 59 SNVFLVEGSEGLIMKVGDFGLRQLVP----LPPDSPFASFVAPEVHKGHPFTDKADMYSI 114
NV L +K+ DFGL ++ L ++ APEV G P+ D++S
Sbjct 146 ENVLLDSQCN---IKIVDFGLSNVMHDGHFLKTSCGSPNYAAPEVISGKPYGPDVDIWSC 202
Query 115 GMLFLGLWLLC 125
G++ LLC
Sbjct 203 GVILYA--LLC 211
> SPBC660.14
Length=581
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 34/119 (28%), Positives = 59/119 (49%), Gaps = 10/119 (8%)
Query 5 SVFVQMEYYELA-LQRLTKEGRTLKIAD---VRRLFMQLLEAIVCVHRAGACHRALHPSN 60
++F+Q++Y E L E L++ D V ++ QL +A+ +H H + PSN
Sbjct 363 NIFLQLDYCENGDLSLFLSELGLLQVMDPFRVWKMLFQLTQALNFIHLLEFVHLDVKPSN 422
Query 61 VFLVEGSEGLIMKVGDFGLRQLVPLPPDSPFAS---FVAPEVHKGHPFTDKADMYSIGM 116
V + +K+GDFGL +P+ ++APE+ H + AD+YS+G+
Sbjct 423 VLITRDGN---LKLGDFGLATSLPVSSMVDLEGDRVYIAPEILASHNYGKPADVYSLGL 478
> Hs9966875
Length=357
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 49/101 (48%), Gaps = 10/101 (9%)
Query 31 DVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVEGSEGLIMKVGDFGLRQL------VP 84
D L Q+L+A+ +HR G HR L P N+ E + + DFGL ++ +
Sbjct 120 DASTLIRQVLDAVYYLHRMGIVHRDLKPENLLYYSQDEESKIMISDFGLSKMEGKGDVMS 179
Query 85 LPPDSPFASFVAPEVHKGHPFTDKADMYSIGMLFLGLWLLC 125
+P +VAPEV P++ D +SIG+ + LLC
Sbjct 180 TACGTP--GYVAPEVLAQKPYSKAVDCWSIGV--IAYILLC 216
> ECU08g1790
Length=309
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 62/124 (50%), Gaps = 6/124 (4%)
Query 6 VFVQMEYYELALQRLTKEGRTLKIADVRRLFMQLLEAIVCVHRAGACHRALHPSNVFLVE 65
+FV +EY L L G L +R + QL+EA+ +H G HR + PSN+ L
Sbjct 75 IFVVLEYMPYDLTGLLASGAKLSTDQIRSITSQLIEAVSSMHGMGLVHRDIKPSNILL-- 132
Query 66 GSEGLIMKVGDFGLRQLVPLPPDSPFAS--FVAPEVHKGH-PFTDKADMYSIGMLFLGLW 122
G+ +K+ DFGL + + + + + APE+ G ++ K D +S+G + L +
Sbjct 133 DCHGM-LKLTDFGLTREISGMMTNRVCTLWYRAPELLLGETSYSLKVDAWSVGCIMLEMR 191
Query 123 LLCP 126
L P
Sbjct 192 LGRP 195
Lambda K H
0.328 0.143 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40