bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
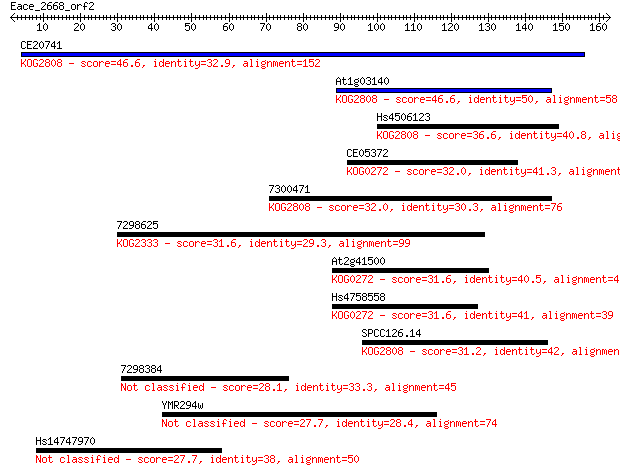
Query= Eace_2668_orf2
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
CE20741 46.6 2e-05
At1g03140 46.6 2e-05
Hs4506123 36.6 0.022
CE05372 32.0 0.51
7300471 32.0 0.57
7298625 31.6 0.67
At2g41500 31.6 0.75
Hs4758558 31.6 0.79
SPCC126.14 31.2 1.0
7298384 28.1 8.5
YMR294w 27.7 9.3
Hs14747970 27.7 9.8
> CE20741
Length=348
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 50/154 (32%), Positives = 77/154 (50%), Gaps = 23/154 (14%)
Query 4 IAKQKQQINEL-PKAGPGKYILQKDLQQKLREQKDREQEETANKKRKTELQQLAELQERL 62
+AK+++ ++ + K G K++ DL+ K ++ +R+Q+E A+KKRK + + L E R
Sbjct 11 MAKKRKAVSGMEVKEGNAKFVKGADLEMKRNQEYERKQQEIASKKRKVDDEILQESSSRT 70
Query 63 KARAFQPHSSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKR 122
K P E E+ + +E P + EI RLR+ PI LFGET
Sbjct 71 K-----PAPVENESEI-----------DEKTP---MSEIQTRLRQRNHPIMLFGETDIDV 111
Query 123 YDRLCKLELQAIDDETTEGQKNVFH-AMQREGEE 155
RL +LEL D EG +N AM+ G+E
Sbjct 112 RKRLHQLELAQPD--LNEGWENELQTAMKVIGKE 143
> At1g03140
Length=420
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 36/61 (59%), Gaps = 5/61 (8%)
Query 89 EEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRL---CKLELQAIDDETTEGQKNV 145
+E L+ P E+ RRLR LKQP++LFGE R DRL K L +D + TEGQ N
Sbjct 99 DENLILPRQ--EVIRRLRFLKQPMTLFGEDDQSRLDRLKYVLKEGLFEVDSDMTEGQTND 156
Query 146 F 146
F
Sbjct 157 F 157
> Hs4506123
Length=342
Score = 36.6 bits (83), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 29/49 (59%), Gaps = 2/49 (4%)
Query 100 EIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDETTEGQKNVFHA 148
E+ RRLR +PI LFGET + + RL K+E+ + E +G +N A
Sbjct 83 EVIRRLRERGEPIRLFGETDYDAFQRLRKIEI--LTPEVNKGLRNDLKA 129
> CE05372
Length=496
Score = 32.0 bits (71), Expect = 0.51, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 92 LVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCKLELQAIDDE 137
L P D ++ +LR L QPI LFGE R +RL L +DE
Sbjct 72 LTLPTDDVQVKLKLRALNQPICLFGEDALDRRERLRALLSTMSEDE 117
> 7300471
Length=340
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 37/76 (48%), Gaps = 5/76 (6%)
Query 71 SSEAEATVADAGDTAEGREEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCKLE 130
S EA+ + + + ++P E+ RRLR +PI +FGET + +DRL + E
Sbjct 52 SVEAQGQTTEGAYSFVADGQNILPRT---EVIRRLRERGEPILIFGETEPEAFDRLRQCE 108
Query 131 LQAIDDETTEGQKNVF 146
+ E G +N F
Sbjct 109 IS--QPEANRGFRNDF 122
> 7298625
Length=604
Score = 31.6 bits (70), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 49/99 (49%), Gaps = 12/99 (12%)
Query 30 QKLREQKDREQEETANKKRKTELQQLAELQERLKARAFQPHSSEAEATVADAGDTAEGRE 89
+KL+EQ+++E+ N +T +Q +E E L A S+ AT D+A GR+
Sbjct 191 RKLKEQQEKEK----NPSEETGEKQPSETTEILDAEKQTDTKSKPTATGCAIDDSAIGRD 246
Query 90 EELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCK 128
+ P VD R + + P++ G P++ R+CK
Sbjct 247 ADHKPAVDF-----REKLVLSPLTTLGNLPFR---RICK 277
> At2g41500
Length=554
Score = 31.6 bits (70), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 88 REEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRLCKL 129
R + P + + RLRRL +PI+LFGE +R RL +L
Sbjct 113 RAAAMAVPTNDKAVRDRLRRLGEPITLFGEQEMERRARLTQL 154
> Hs4758558
Length=522
Score = 31.6 bits (70), Expect = 0.79, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 88 REEELVPPVDLPEIFRRLRRLKQPISLFGETPWKRYDRL 126
R ++ D E+ LR L +PI+LFGE P +R +RL
Sbjct 95 RARQINVSTDDSEVKACLRALGEPITLFGEGPAERRERL 133
> SPCC126.14
Length=343
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 34/55 (61%), Gaps = 5/55 (9%)
Query 96 VDLPEIFRRLRRLKQPISLFGET---PWKRYDRLCKL-ELQAIDDE-TTEGQKNV 145
+ L EI +LR +K+PI LFGE+ +RY L K +L+ I++E T+G + +
Sbjct 124 LTLTEIIAKLREMKEPIRLFGESEEATIQRYYSLLKYKKLEEIENELLTKGVETI 178
> 7298384
Length=499
Score = 28.1 bits (61), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Query 31 KLREQKDREQEETANKKRKTELQQLAELQERLKARAFQPHSSEAE 75
++R +K++ EE A K R +++ L E+Q++LK+ FQ H + +
Sbjct 288 QIRNEKEKLAEEVATKDR--DIENLQEMQQQLKSELFQVHGQQGQ 330
> YMR294w
Length=373
Score = 27.7 bits (60), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 5/75 (6%)
Query 42 ETANKKRKTELQQLAELQERLKARAFQPHSSEAEATVADAGDTAEGREEELVPPVDL-PE 100
E + +TE+++L E+Q +L + SS +T EG++ ++P + L
Sbjct 116 ENLTSEMQTEIKELCEIQSKLATES----SSRLTNLRKKLLETYEGQDTVILPNIILDTS 171
Query 101 IFRRLRRLKQPISLF 115
+RL++L Q ISL
Sbjct 172 NIKRLQKLDQKISLM 186
> Hs14747970
Length=794
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 9/50 (18%)
Query 8 KQQINELPKAGPGKYILQKDLQQKLREQKDREQEETANKKRKTELQQLAE 57
KQ +++ K G G Y L +L D+EQE+ A+ RK ELQ+L+
Sbjct 737 KQLVSQSKKTGQGDYPLNNEL--------DKEQEDVASTTRK-ELQELSS 777
Lambda K H
0.311 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40