bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2690_orf1
Length=120
Score E
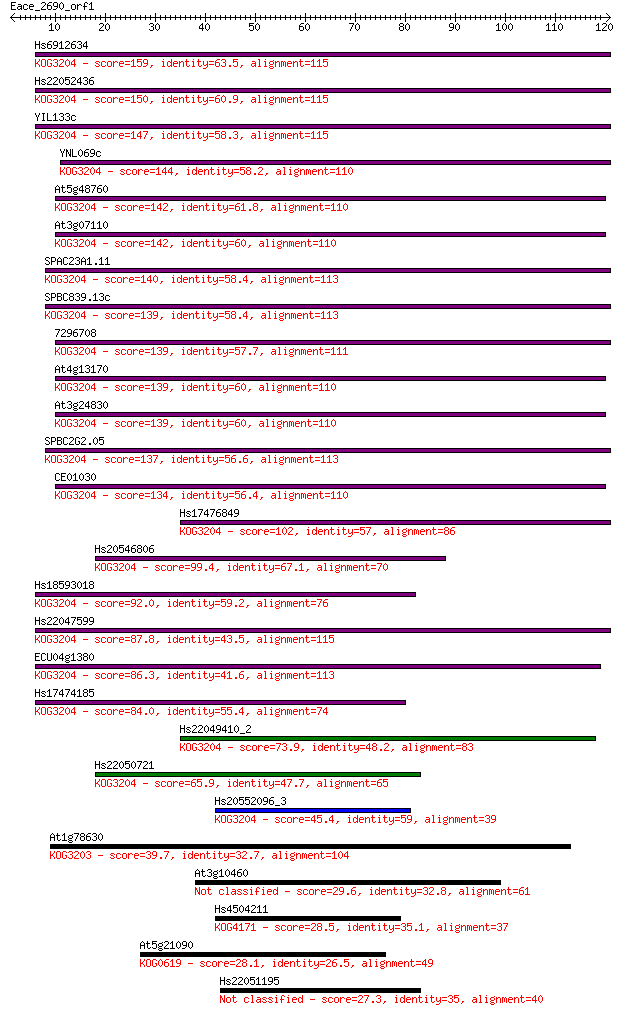
Sequences producing significant alignments: (Bits) Value
Hs6912634 159 1e-39
Hs22052436 150 6e-37
YIL133c 147 5e-36
YNL069c 144 4e-35
At5g48760 142 2e-34
At3g07110 142 2e-34
SPAC23A1.11 140 6e-34
SPBC839.13c 139 8e-34
7296708 139 1e-33
At4g13170 139 1e-33
At3g24830 139 1e-33
SPBC2G2.05 137 4e-33
CE01030 134 6e-32
Hs17476849 102 2e-22
Hs20546806 99.4 2e-21
Hs18593018 92.0 2e-19
Hs22047599 87.8 4e-18
ECU04g1380 86.3 1e-17
Hs17474185 84.0 6e-17
Hs22049410_2 73.9 6e-14
Hs22050721 65.9 2e-11
Hs20552096_3 45.4 3e-05
At1g78630 39.7 0.001
At3g10460 29.6 1.5
Hs4504211 28.5 2.9
At5g21090 28.1 4.3
Hs22051195 27.3 6.7
> Hs6912634
Length=203
Score = 159 bits (402), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 73/115 (63%), Positives = 94/115 (81%), Gaps = 0/115 (0%)
Query 6 MAFKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRL 65
MA +V+V++ +GHLLGRLA+ VAK++ G+ VV V+CE INISG+ +RNKLKY AFLR
Sbjct 1 MAEVQVLVLDGRGHLLGRLAAIVAKQVLLGRKVVVVRCEGINISGNFYRNKLKYLAFLRK 60
Query 66 RMNSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
RMN+NP RGPYH RAPSRIFWR VRGM+ HKT RG+ AL RL+V++G+P Y++K
Sbjct 61 RMNTNPSRGPYHFRAPSRIFWRTVRGMLPHKTKRGQAALDRLKVFDGIPPPYDKK 115
> Hs22052436
Length=203
Score = 150 bits (379), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 70/115 (60%), Positives = 91/115 (79%), Gaps = 0/115 (0%)
Query 6 MAFKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRL 65
MA +V+V++ +GHLLGRLA+ VAK++ G+ VV V CE INISG+ +RNKL Y AFLR
Sbjct 1 MAEVQVLVLDGRGHLLGRLAAIVAKQVLLGRKVVVVCCEGINISGNFYRNKLNYLAFLRK 60
Query 66 RMNSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
RMN++P RGPYH RAPSR+FWR VRGM+ HKT RG+ AL RL+V +G+P Y++K
Sbjct 61 RMNTSPSRGPYHFRAPSRVFWRTVRGMLPHKTKRGQAALDRLKVSDGIPPPYDKK 115
> YIL133c
Length=199
Score = 147 bits (371), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 67/115 (58%), Positives = 87/115 (75%), Gaps = 0/115 (0%)
Query 6 MAFKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRL 65
M+ + VVVI+ +GHL+GRLAS VAK++ NGQ +V V+ E +NISG RNKLKY FLR
Sbjct 1 MSVEPVVVIDGKGHLVGRLASVVAKQLLNGQKIVVVRAEELNISGEFFRNKLKYHDFLRK 60
Query 66 RMNSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
N RGP+H RAPSRIF++A+RGM+ HKT RGK AL RL+V+EG+P Y++K
Sbjct 61 ATAFNKTRGPFHFRAPSRIFYKALRGMVSHKTARGKAALERLKVFEGIPPPYDKK 115
> YNL069c
Length=198
Score = 144 bits (363), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 64/110 (58%), Positives = 83/110 (75%), Gaps = 0/110 (0%)
Query 11 VVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNSN 70
VVVI+ + HLLGRLAST+AK++ NGQ +V V+ E +NISG RNKLKY FLR N
Sbjct 5 VVVIDAKDHLLGRLASTIAKQVLNGQKIVVVRAEALNISGEFFRNKLKYHDFLRKATAFN 64
Query 71 PRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
RGP+H RAPSRI ++A+RGM+ HKT RGK AL RL+++EG+P Y++K
Sbjct 65 KTRGPFHFRAPSRILYKAIRGMVSHKTARGKAALERLKIFEGIPPPYDKK 114
> At5g48760
Length=206
Score = 142 bits (358), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 68/110 (61%), Positives = 82/110 (74%), Gaps = 0/110 (0%)
Query 10 KVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNS 69
K VV++ + H+LGRLAS AKE+ NGQ VV V+CE I +SG L R K+KY FLR RMN+
Sbjct 10 KRVVVDARHHMLGRLASITAKELLNGQKVVIVRCEEICLSGGLVRQKMKYMRFLRKRMNT 69
Query 70 NPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYER 119
P GP H RAPS+IFWR VRGMI HKT RG AL+RL+VYEGVP Y++
Sbjct 70 KPSHGPIHFRAPSKIFWRTVRGMIPHKTKRGAAALARLKVYEGVPTPYDK 119
> At3g07110
Length=206
Score = 142 bits (357), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 66/110 (60%), Positives = 84/110 (76%), Gaps = 0/110 (0%)
Query 10 KVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNS 69
K VV++ + H+LGRLAS VAK++ NGQ++V V+CE I +SG L R K+KY FLR RMN+
Sbjct 10 KRVVVDARHHMLGRLASVVAKDLLNGQNIVVVRCEEICLSGGLVRQKMKYMRFLRKRMNT 69
Query 70 NPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYER 119
P GP H RAPS+IFWR VRGMI HKT RG AL+RL+V+EGVP Y++
Sbjct 70 KPSHGPIHFRAPSKIFWRTVRGMIPHKTKRGANALARLKVFEGVPTPYDK 119
> SPAC23A1.11
Length=197
Score = 140 bits (353), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 66/113 (58%), Positives = 84/113 (74%), Gaps = 0/113 (0%)
Query 8 FKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRM 67
F+KVVVI+ +GHLLGRLAS VAK++ GQ VV V+CE +NISG RNKLKY A+LR
Sbjct 4 FQKVVVIDAKGHLLGRLASVVAKQLLGGQKVVVVRCEELNISGHFFRNKLKYLAYLRKAC 63
Query 68 NSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
NP RG +H RAPSRIF +AVRGM+ HKT RG+ AL L+ EG+P ++++
Sbjct 64 RYNPSRGAFHFRAPSRIFQKAVRGMLPHKTARGQAALEHLQAVEGIPPPFDKQ 116
> SPBC839.13c
Length=197
Score = 139 bits (351), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 66/113 (58%), Positives = 84/113 (74%), Gaps = 0/113 (0%)
Query 8 FKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRM 67
F+KVVVI+ +GHLLGRLAS VAK++ GQ VV V+CE +NISG RNKLKY A+LR
Sbjct 4 FQKVVVIDAKGHLLGRLASVVAKQLLGGQKVVVVRCEELNISGHFFRNKLKYLAYLRKAC 63
Query 68 NSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
NP RG +H RAPSRIF +AVRGM+ HKT RG+ AL L+ EG+P ++++
Sbjct 64 RYNPSRGAFHFRAPSRIFQKAVRGMLPHKTARGQAALEHLQAVEGIPPPFDKQ 116
> 7296708
Length=205
Score = 139 bits (351), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 64/111 (57%), Positives = 86/111 (77%), Gaps = 0/111 (0%)
Query 10 KVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNS 69
+ VVI+ +GHLLGRLAS VAK + G V V+CE +N+SG +RNK+K+ A+LR R N
Sbjct 7 RTVVIDGRGHLLGRLASVVAKYLLQGGKVAVVRCEELNLSGHFYRNKIKFLAYLRKRCNV 66
Query 70 NPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
NP RGP+H RAPSRIF++AVRGMI HKT RG+ AL+RL V++G+P Y+++
Sbjct 67 NPARGPFHFRAPSRIFYKAVRGMIPHKTKRGQAALARLRVFDGIPSPYDKR 117
> At4g13170
Length=206
Score = 139 bits (350), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 66/110 (60%), Positives = 82/110 (74%), Gaps = 0/110 (0%)
Query 10 KVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNS 69
K VV++ + H+LGRLAS AKE+ NGQ VV V+CE I +SG L R K+KY FLR RMN+
Sbjct 10 KRVVVDGRHHMLGRLASNTAKELLNGQEVVVVRCEEICLSGGLVRQKMKYMRFLRKRMNT 69
Query 70 NPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYER 119
P GP H RAPS+IFWR VRGMI HKT RG AL+RL+V+EG+P Y++
Sbjct 70 KPSHGPIHFRAPSKIFWRTVRGMIPHKTKRGAAALARLKVFEGIPPPYDK 119
> At3g24830
Length=206
Score = 139 bits (350), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 66/110 (60%), Positives = 82/110 (74%), Gaps = 0/110 (0%)
Query 10 KVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNS 69
K VV++ + H+ GRLAS +AKE+ NGQ VV V+CE I +SG L R K+KY FLR RMN+
Sbjct 10 KRVVVDARHHMCGRLASIIAKELLNGQSVVVVRCEEICLSGGLVRQKMKYMRFLRKRMNT 69
Query 70 NPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYER 119
P GP H RAPS+IFWR VRGMI HKT RG AL+RL+V+EGVP Y++
Sbjct 70 KPSHGPIHFRAPSKIFWRTVRGMIPHKTKRGAAALARLKVFEGVPPPYDK 119
> SPBC2G2.05
Length=197
Score = 137 bits (346), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 64/113 (56%), Positives = 84/113 (74%), Gaps = 0/113 (0%)
Query 8 FKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRM 67
F+K+V+I+ +GHL+GRLASTVAK++ GQ VV V+CE +NISG RNKLKY A+LR
Sbjct 4 FQKLVIIDAKGHLMGRLASTVAKQLLAGQKVVVVRCEELNISGHFFRNKLKYLAYLRKAC 63
Query 68 NSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
NP RG +H RAPSRIF +AVRGM+ HKT RG AL L+ EG+P ++++
Sbjct 64 RYNPSRGAFHFRAPSRIFTKAVRGMLPHKTTRGNIALKNLQALEGIPPPFDKQ 116
> CE01030
Length=202
Score = 134 bits (336), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 62/110 (56%), Positives = 80/110 (72%), Gaps = 0/110 (0%)
Query 10 KVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNS 69
+ ++I+ + HLLGRLAS VAK++ G VV ++ E I ISG+ HR+KLKY +FLR R N
Sbjct 6 RAIIIDGKNHLLGRLASIVAKKLLQGDKVVVLRAEEIVISGNFHRSKLKYMSFLRKRCNI 65
Query 70 NPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYER 119
NP RG +H RAP +IFWR VRGM+ HKT RG +AL L YEGVP KY++
Sbjct 66 NPARGAFHYRAPGKIFWRTVRGMLPHKTNRGNEALKNLRAYEGVPAKYQK 115
> Hs17476849
Length=359
Score = 102 bits (253), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 49/87 (56%), Positives = 62/87 (71%), Gaps = 1/87 (1%)
Query 35 GQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNSNPRRGPYHMRAPSRIFWRAVRG-MI 93
G+ VV V+CE INISG+ +RNKLK AFL RMN+NP R YH RAPSRIFW M+
Sbjct 195 GRKVVVVRCEGINISGNFYRNKLKNLAFLGKRMNTNPFRSAYHFRAPSRIFWPTCEAYML 254
Query 94 RHKTIRGKKALSRLEVYEGVPVKYERK 120
RHKT RG +L L+V++G+P Y++K
Sbjct 255 RHKTKRGHASLDCLKVFDGIPPPYDKK 281
> Hs20546806
Length=303
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/70 (67%), Positives = 53/70 (75%), Gaps = 0/70 (0%)
Query 18 GHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNSNPRRGPYH 77
GHLLGRLA+ VAK++ G+ V V CE INISGS RNKLKY AFL RMN+NP R PYH
Sbjct 68 GHLLGRLAAIVAKQVLLGRKAVVVSCEGINISGSFCRNKLKYLAFLHKRMNTNPSRRPYH 127
Query 78 MRAPSRIFWR 87
+ PSRIFWR
Sbjct 128 FQVPSRIFWR 137
> Hs18593018
Length=105
Score = 92.0 bits (227), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 45/76 (59%), Positives = 59/76 (77%), Gaps = 0/76 (0%)
Query 6 MAFKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRL 65
MA +V+V++ +GHLL RLA+ VAK++ G+ VV V+CE INISG+ +RNKLKY FLR
Sbjct 1 MAEVQVLVLDGRGHLLVRLAAIVAKQVLLGRKVVVVRCEGINISGNFYRNKLKYLGFLRK 60
Query 66 RMNSNPRRGPYHMRAP 81
RMN++ RGPYH RAP
Sbjct 61 RMNTHLSRGPYHFRAP 76
> Hs22047599
Length=162
Score = 87.8 bits (216), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 50/115 (43%), Positives = 68/115 (59%), Gaps = 17/115 (14%)
Query 6 MAFKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRL 65
MA +V V++ +GHLLGRLA+ VAK++ G VV V+CE INISG+ +RNKL
Sbjct 1 MAEVQVPVLHGRGHLLGRLAAIVAKQVMLGWKVVVVRCEGINISGNFYRNKLNCS----- 55
Query 66 RMNSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYERK 120
R PS IF VRGM+ HKT RG+ L L+V++G+ Y++K
Sbjct 56 ------------FRTPSCIFRWTVRGMLPHKTKRGQAVLDHLQVFDGISPLYDKK 98
> ECU04g1380
Length=198
Score = 86.3 bits (212), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/113 (41%), Positives = 64/113 (56%), Gaps = 0/113 (0%)
Query 6 MAFKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRL 65
M K+ V+I+ G + G+LA+ VAK + G V V E GSL R K++ +L
Sbjct 1 MEKKEHVIIDATGQVAGKLAAKVAKLLLEGVRVTVVCAEEAVFVGSLERAMDKFKQYLNK 60
Query 66 RMNSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEGVPVKYE 118
R NPRRGP+H R P + +R MI +K RGK A+SRL YEG+P + E
Sbjct 61 RCLVNPRRGPFHFREPRMHLAKIIRRMISYKKPRGKAAMSRLATYEGIPRELE 113
> Hs17474185
Length=150
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 41/74 (55%), Positives = 56/74 (75%), Gaps = 0/74 (0%)
Query 6 MAFKKVVVINCQGHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRL 65
MA +V+V++ +GHLLG LA+ VAK++ G+ VV V+CE I+ISG+ + NKLKY AFLR
Sbjct 1 MAEGQVLVLDGRGHLLGCLAAIVAKQVLLGRKVVIVRCEGIDISGNFYGNKLKYLAFLRK 60
Query 66 RMNSNPRRGPYHMR 79
RMN++P RGP R
Sbjct 61 RMNTSPSRGPTTCR 74
> Hs22049410_2
Length=176
Score = 73.9 bits (180), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 40/86 (46%), Positives = 53/86 (61%), Gaps = 3/86 (3%)
Query 35 GQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNSNPRRGPYHMRAPSRIFWRAVRGMIR 94
G VV + E I ISG+ +RNKLKY AFL MN+NP RGP+H RA S +F + RGM+
Sbjct 4 GWKVVVMYGEGIGISGNFYRNKLKYLAFLCKWMNTNPSRGPHHPRAASLLFRQTPRGMLP 63
Query 95 HKTIRGKKALS---RLEVYEGVPVKY 117
HKT RG+ + L + G+ + Y
Sbjct 64 HKTKRGQPPWTASCHLPILLGISMGY 89
> Hs22050721
Length=188
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 45/65 (69%), Gaps = 1/65 (1%)
Query 18 GHLLGRLASTVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNSNPRRGPYH 77
G+LLG LA+ V K++ G+ VV V+CE++N+S + +RNKLKY AF R RM++NP P
Sbjct 75 GYLLGHLAAVVVKQVLLGRKVV-VRCESVNVSDNFYRNKLKYPAFFRRRMSTNPDEAPVI 133
Query 78 MRAPS 82
P+
Sbjct 134 SWPPA 138
> Hs20552096_3
Length=139
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/39 (58%), Positives = 27/39 (69%), Gaps = 2/39 (5%)
Query 42 KCENINISGSLHRNKLKYQAFLRLRMNSNPRRGPYHMRA 80
+CE+INISG +RNKLKY FLR RMN+ R P H A
Sbjct 1 QCESINISGKFYRNKLKYLGFLRKRMNTFWR--PCHFSA 37
> At1g78630
Length=241
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 50/116 (43%), Gaps = 13/116 (11%)
Query 9 KKVVVINCQGHLLGRLASTVAKEIQN------------GQHVVCVKCENINISGSLHRNK 56
K V++ +LGRLAST+A I+ G V+ V E + +SG K
Sbjct 103 KPWFVVDATDKILGRLASTIANHIRGKNLASYTPSVDMGAFVIVVNAEKVAVSGKKRNQK 162
Query 57 LKYQAFLRLRMNSNPRRGPYHMRAPSRIFWRAVRGMIRHKTIRGKKALSRLEVYEG 112
L + R + R P RI AVRGM+ + G+ + L+VY+G
Sbjct 163 LYRRHSGRPGGMTVETFDQLQQRIPERIVEHAVRGMLPKGRL-GRALFNHLKVYKG 217
> At3g10460
Length=132
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 38 VVCVKCENINISGSLH-RNKLKYQAFLRLRMNSNPRRGPYHMRAPSRIFWRAVRGMIRHK 96
VVC+ EN+ I GS+ N L + +L+++ S +H+R P R + A I K
Sbjct 11 VVCLYTENV-IGGSVFIYNNLPHGHYLQVKCKSGDTNLGFHVRRPGRFYNYAFTDHILGK 69
Query 97 TI 98
T+
Sbjct 70 TL 71
> Hs4504211
Length=732
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 42 KCENINISGSLHRNKLKYQAFLRLRMNSNPRRGPYHM 78
+CEN NI +L + + A LR+ +N+ R P+H+
Sbjct 295 ECENTNIMKNLPQGTSQVPADLRISINTFCRAFPFHL 331
> At5g21090
Length=218
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 27 TVAKEIQNGQHVVCVKCENINISGSLHRNKLKYQAFLRLRMNSNPRRGP 75
T+ E+ N ++++ + N N++G + + K ++ + LR+N N GP
Sbjct 109 TIPSELGNLKNLISLDLYNNNLTGIVPTSLGKLKSLVFLRLNDNRLTGP 157
> Hs22051195
Length=784
Score = 27.3 bits (59), Expect = 6.7, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 20/41 (48%), Gaps = 1/41 (2%)
Query 43 CENINIS-GSLHRNKLKYQAFLRLRMNSNPRRGPYHMRAPS 82
C ++N G LH NK Q L++ P P ++APS
Sbjct 7 CVSLNTGMGELHENKCMLQFLCELQLLDQPIAFPQSLKAPS 47
Lambda K H
0.325 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40