bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
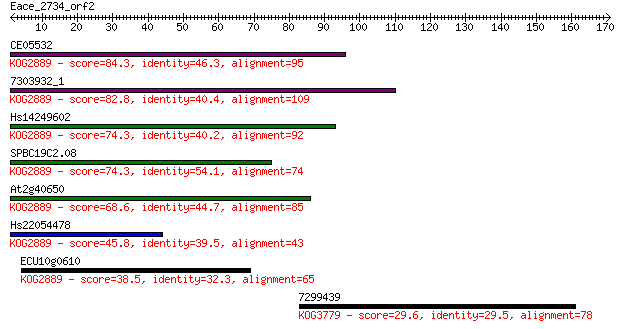
Query= Eace_2734_orf2
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
CE05532 84.3 1e-16
7303932_1 82.8 3e-16
Hs14249602 74.3 1e-13
SPBC19C2.08 74.3 1e-13
At2g40650 68.6 6e-12
Hs22054478 45.8 4e-05
ECU10g0610 38.5 0.007
7299439 29.6 3.0
> CE05532
Length=320
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 44/95 (46%), Positives = 60/95 (63%), Gaps = 1/95 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
+FKY+RA+ YLRL S E+Y +LEPL D+RKLR + G F YMD F+DN LR+
Sbjct 95 EFKYIRALGAMYLRLTFDSTEIYKYLEPLYNDFRKLRYMNKMGRFEAIYMDDFIDNLLRE 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALDVEV 95
+ DI LP + KR AL + +LP + S LD ++
Sbjct 155 DRYCDIQLPRLQKRWALEEVDMLPSYK-SLLDGDL 188
> 7303932_1
Length=355
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 44/109 (40%), Positives = 68/109 (62%), Gaps = 4/109 (3%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
+FKY+RA+ FYLRL A+ + Y +LEPL D RKLR ++ +G F + YMD ++D LR
Sbjct 95 EFKYVRALGAFYLRLTGAALDCYKYLEPLYIDNRKLRRQNRAGQFEIVYMDEYIDELLRN 154
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALDVEVSEQEKSLPLTDSQA 109
+ + DI LP I KR L + + P ++S LD ++ ++ LP + +A
Sbjct 155 DRVCDIILPRIQKRSILEENNEIEP-KVSVLDEDLDDE---LPSDEEKA 199
> Hs14249602
Length=236
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 57/92 (61%), Gaps = 1/92 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
DFKY+R + Y+RL + + Y +LEPL DYRK++ ++ +G F L ++D F+D L
Sbjct 19 DFKYVRMLGALYMRLTGTAIDCYKYLEPLYNDYRKIKSQNRNGEFELMHVDEFIDELLHS 78
Query 61 NTLFDIDLPPIVKRKALVQQQLLPPLRLSALD 92
+ DI LP + KR L + + L P R+SAL+
Sbjct 79 ERVCDIILPRLQKRYVLEEAEQLEP-RVSALE 109
> SPBC19C2.08
Length=210
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/74 (54%), Positives = 49/74 (66%), Gaps = 1/74 (1%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRK 60
+FKYLRA+A FY+RL EV+ LEPLL DYRKLR+R NS LTY+D VD+ L
Sbjct 98 EFKYLRALAAFYVRLTWDDVEVHQTLEPLLRDYRKLRIRTNS-EIRLTYLDEVVDDLLNA 156
Query 61 NTLFDIDLPPIVKR 74
+ DI LPP+ R
Sbjct 157 EVVCDISLPPLRSR 170
> At2g40650
Length=363
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 38/93 (40%), Positives = 53/93 (56%), Gaps = 8/93 (8%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGA--------FSLTYMDV 52
D+KY+R + FYLRL +VY +LEPL DYRK+R + + G FSLT++D
Sbjct 95 DYKYVRILGAFYLRLTGTDVDVYRYLEPLYNDYRKVRQKLSDGNLFLWFGIEFSLTHVDE 154
Query 53 FVDNCLRKNTLFDIDLPPIVKRKALVQQQLLPP 85
++ L K+ DI +P + KR L Q LL P
Sbjct 155 VIEELLTKDYSCDIAMPRLKKRWTLEQNGLLEP 187
> Hs22054478
Length=264
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 1 DFKYLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSG 43
DFKY+ + Y+RL+ + + Y +LEPL DYRK++ ++ +G
Sbjct 95 DFKYVHMLGALYMRLMGTAIDCYKYLEPLYNDYRKIKSQNRNG 137
> ECU10g0610
Length=164
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 4 YLRAVAVFYLRLVAASDEVYIHLEPLLTDYRKLRVRDNSGAFSLTYMDVFVDNCLRKNTL 63
Y A ++FYLRL + + L D+RK+ V D + Y+DV D+ L K+ +
Sbjct 96 YFIAASLFYLRLSKRFHKYRPLIGLFLADFRKIPVVDGQNNRTFMYLDVLADDLLNKSRI 155
Query 64 FDIDL 68
F++ L
Sbjct 156 FNVHL 160
> 7299439
Length=1703
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query 83 LPPLRLSALDVEVSEQEK-SLPLTDSQAHALLLSKRLLQQQQHKLQQHVLQQRNRQLQQQ 141
L + L D E +E E + + +AL R +Q+Q ++QQ +Q +R ++Q+
Sbjct 816 LQSMMLDQEDSESNELESPQIQQKRVEKNALKSQLRSMQEQLAEMQQKYVQLCSR-MEQE 874
Query 142 KEIEEERQMQLIKEETAPD 160
E +E Q Q +++E PD
Sbjct 875 SECQELDQDQDVEQEQEPD 893
Lambda K H
0.321 0.136 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2490594054
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40