bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2761_orf1
Length=134
Score E
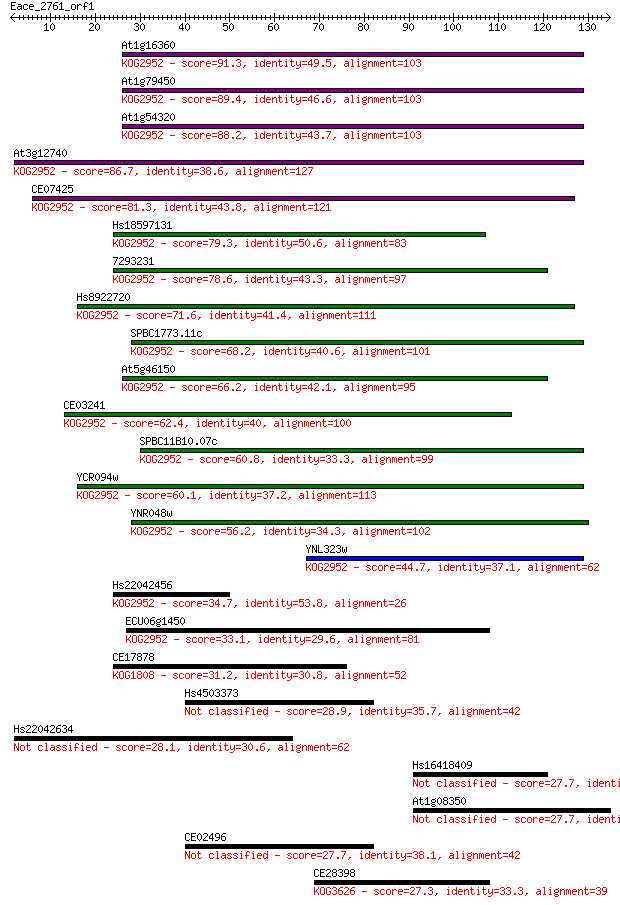
Sequences producing significant alignments: (Bits) Value
At1g16360 91.3 5e-19
At1g79450 89.4 2e-18
At1g54320 88.2 4e-18
At3g12740 86.7 1e-17
CE07425 81.3 4e-16
Hs18597131 79.3 2e-15
7293231 78.6 3e-15
Hs8922720 71.6 4e-13
SPBC1773.11c 68.2 4e-12
At5g46150 66.2 1e-11
CE03241 62.4 2e-10
SPBC11B10.07c 60.8 6e-10
YCR094w 60.1 1e-09
YNR048w 56.2 2e-08
YNL323w 44.7 5e-05
Hs22042456 34.7 0.051
ECU06g1450 33.1 0.16
CE17878 31.2 0.64
Hs4503373 28.9 2.6
Hs22042634 28.1 4.5
Hs16418409 27.7 5.8
At1g08350 27.7 6.5
CE02496 27.7 6.8
CE28398 27.3 8.0
> At1g16360
Length=316
Score = 91.3 bits (225), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 51/107 (47%), Positives = 63/107 (58%), Gaps = 8/107 (7%)
Query 26 ENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVT-NNTYDVKSFGGSKSVVVTQAS 84
E IVWMR A LP FRK+Y + + L+ +KV N Y+ SF G K +V++ S
Sbjct 204 EQEDLIVWMRTAALPTFRKLYGK-IDTDLQAGDTIKVLLQNNYNTYSFNGKKKLVLSTTS 262
Query 85 WLGGRTSFLGIAYLVVGSVCLLF---FGVLFYYHRRNPRHLGDVSWL 128
WLGGR FLGIAYL VGS+CL F VL+ PR LGD S+L
Sbjct 263 WLGGRNDFLGIAYLTVGSICLFLAVSFSVLYLAK---PRQLGDPSYL 306
> At1g79450
Length=350
Score = 89.4 bits (220), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/106 (45%), Positives = 58/106 (54%), Gaps = 6/106 (5%)
Query 26 ENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSFGGSKSVVVTQASW 85
E IVWMR A LP FRK+Y + + N Y+ SF G K +V++ SW
Sbjct 238 EQEDLIVWMRTAALPTFRKLYGKIETDLHAGDTITVLLQNNYNTYSFNGQKKLVLSTTSW 297
Query 86 LGGRTSFLGIAYLVVGSVCLLF---FGVLFYYHRRNPRHLGDVSWL 128
LGGR FLGIAYL VGS+CL F VL+ PR LGD S+L
Sbjct 298 LGGRNDFLGIAYLTVGSICLFLAVTFAVLYLVK---PRQLGDPSYL 340
> At1g54320
Length=349
Score = 88.2 bits (217), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 45/104 (43%), Positives = 60/104 (57%), Gaps = 2/104 (1%)
Query 26 ENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVT-NNTYDVKSFGGSKSVVVTQAS 84
E IVWMR A LP FRK+Y + + L++ + V NN Y+ SF G K +V++ S
Sbjct 238 EQEDLIVWMRTAALPTFRKLYGK-IESDLEMGDTIHVKLNNNYNTYSFNGKKKLVLSTTS 296
Query 85 WLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHRRNPRHLGDVSWL 128
WLGG+ FLGIAYL VG +C + + PR LGD S+L
Sbjct 297 WLGGKNDFLGIAYLTVGGICFILALAFTIMYLVKPRRLGDPSYL 340
> At3g12740
Length=350
Score = 86.7 bits (213), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 49/128 (38%), Positives = 70/128 (54%), Gaps = 6/128 (4%)
Query 2 DFWLFNQTYASALHMDKPGVGWGVENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLK 61
+F N T ++L +KP + IVWMR A LP FRK+Y + + L+ ++
Sbjct 219 NFQKGNLTGGASLDPNKPLS----DQEDLIVWMRTAALPTFRKLYGK-IESDLEKGENIQ 273
Query 62 VT-NNTYDVKSFGGSKSVVVTQASWLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHRRNPR 120
VT N Y+ SF G K +V++ SWLGG+ FLGIAYL VG +C + + PR
Sbjct 274 VTLQNNYNTYSFSGKKKLVLSTTSWLGGKNDFLGIAYLTVGGICFVLALAFTVMYLVKPR 333
Query 121 HLGDVSWL 128
LGD ++L
Sbjct 334 RLGDPTYL 341
> CE07425
Length=348
Score = 81.3 bits (199), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 53/129 (41%), Positives = 68/129 (52%), Gaps = 9/129 (6%)
Query 6 FNQTYASALHMDKPGVGWGVENSHFIVWMREAPLPVFRKVYARFV---AQPL---KLP-- 57
FN T P G EN FIVWMR A LP F+K++ R V PL LP
Sbjct 214 FNNTTKPPNWSKNPCEVGGFENVDFIVWMRTAALPYFKKLW-RIVDRTTNPLFSNGLPQG 272
Query 58 FYLKVTNNTYDVKSFGGSKSVVVTQASWLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHRR 117
Y+ N Y V+SFGG K V++ SW GG+ SFLGIAYLVVGS+ ++ V H +
Sbjct 273 TYILTVENNYPVQSFGGKKEFVISTTSWAGGKNSFLGIAYLVVGSLAIVLGVVFIVIHMK 332
Query 118 NPRHLGDVS 126
+ ++S
Sbjct 333 FGHSMNELS 341
> Hs18597131
Length=351
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 42/88 (47%), Positives = 58/88 (65%), Gaps = 6/88 (6%)
Query 24 GVENSHFIVWMREAPLPVFRKVYAR-----FVAQPLKLPFYLKVTNNTYDVKSFGGSKSV 78
G N F+VWMR A LP FRK+YAR + A + + + +T N Y V++FGG K +
Sbjct 244 GFINQDFVVWMRTAALPTFRKLYARIRQGNYSAGLPRGAYRVNITYN-YPVRAFGGHKLL 302
Query 79 VVTQASWLGGRTSFLGIAYLVVGSVCLL 106
+ + SW+GG+ FLGIAYLVVGS+C+L
Sbjct 303 IFSSISWMGGKNPFLGIAYLVVGSLCIL 330
> 7293231
Length=357
Score = 78.6 bits (192), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 42/103 (40%), Positives = 57/103 (55%), Gaps = 6/103 (5%)
Query 24 GVENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYDVK------SFGGSKS 77
G +N IVWMR A LP FRK+Y R LK N T ++K SF G+K
Sbjct 240 GFQNEDLIVWMRTAALPSFRKLYRRLNQTNTNYANGLKSGNYTLNIKYNYPVVSFDGTKR 299
Query 78 VVVTQASWLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHRRNPR 120
++++ S LGG+ FLGIAY+VVG++C+ L + H R R
Sbjct 300 MILSTTSVLGGKNPFLGIAYIVVGAICITLGLALLFIHMRCSR 342
> Hs8922720
Length=361
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 46/120 (38%), Positives = 63/120 (52%), Gaps = 11/120 (9%)
Query 16 MDKPGVGWGVENSHFIVWMREAPLPVFRKVYARFVAQPLKL-------PFYLKVTNNTYD 68
+D G N FIVWMR A LP FRK+Y R + + L + L VT N Y
Sbjct 243 LDSDPDNNGFINEDFIVWMRTAALPTFRKLY-RLIERKSDLHPTLPAGRYSLNVTYN-YP 300
Query 69 VKSFGGSKSVVVTQASWLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHR--RNPRHLGDVS 126
V F G K ++++ SW+GG+ FLGIAY+ VGS+ L VL + RN + D++
Sbjct 301 VHYFDGRKRMILSTISWMGGKNPFLGIAYIAVGSISFLLGVVLLVINHKYRNSSNTADIT 360
> SPBC1773.11c
Length=396
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 41/101 (40%), Positives = 54/101 (53%), Gaps = 0/101 (0%)
Query 28 SHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSFGGSKSVVVTQASWLG 87
+ VWMR A LP F K+ R + Y + VKSF G+KS+V+T S LG
Sbjct 279 ENLQVWMRTAGLPTFSKLAMRNDNDDIFPGTYEIKIGLFFPVKSFDGTKSLVLTTRSVLG 338
Query 88 GRTSFLGIAYLVVGSVCLLFFGVLFYYHRRNPRHLGDVSWL 128
G+ FLGIAY+VV +VC++ V H PR L D +L
Sbjct 339 GKNPFLGIAYIVVSAVCVVLGTVFTLRHFIRPRKLADHRYL 379
> At5g46150
Length=329
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 40/96 (41%), Positives = 58/96 (60%), Gaps = 2/96 (2%)
Query 26 ENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVT-NNTYDVKSFGGSKSVVVTQAS 84
+ FIVWMR A L FRK+Y R + + L+ ++V N Y+ SF G K ++++ ++
Sbjct 235 DQEDFIVWMRAAALLSFRKLYGR-IEEDLEPGKVVEVNLMNNYNTYSFSGQKKLILSTSN 293
Query 85 WLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHRRNPR 120
WLGGR FLGI YLVVGS ++ + H +NPR
Sbjct 294 WLGGRNDFLGITYLVVGSSSIVISIIFMLLHLKNPR 329
> CE03241
Length=361
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/106 (37%), Positives = 52/106 (49%), Gaps = 7/106 (6%)
Query 13 ALHMDKPGVGWGVENSHFIVWMREAPLPVFRKVY------ARFVAQPLKLPFYLKVTNNT 66
L+ P VG G EN F+VWM+ A LP FRK+Y + L Y N
Sbjct 242 GLNSIDPDVGIGFENIDFMVWMKVAALPKFRKLYRILNRQVDMFSNGLPKGQYQLTINYN 301
Query 67 YDVKSFGGSKSVVVTQASWLGGRTSFLGIAYLVVGSVCLLFFGVLF 112
Y V + G K V+ +W+G R FL + YLVVG+ LL +LF
Sbjct 302 YPVDMYSGDKYFVIANENWVGPRNLFLPVIYLVVGTF-LLLVTILF 346
> SPBC11B10.07c
Length=371
Score = 60.8 bits (146), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 33/99 (33%), Positives = 50/99 (50%), Gaps = 0/99 (0%)
Query 30 FIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSFGGSKSVVVTQASWLGGR 89
F +WMR A LP F K+ R V L+ Y + V + G+K+++ + S +GG+
Sbjct 265 FQIWMRAAALPTFSKLALRNVTTALQPGIYEMNITYNFPVTEYKGTKTIMFSTTSVIGGK 324
Query 90 TSFLGIAYLVVGSVCLLFFGVLFYYHRRNPRHLGDVSWL 128
FLGI Y V+G +C +L PR +GD +L
Sbjct 325 NYFLGILYFVIGGLCAASGVILSIACLIKPRRVGDPRYL 363
> YCR094w
Length=391
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 58/121 (47%), Gaps = 9/121 (7%)
Query 16 MDKPGVGWGVEN-------SHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYD 68
M K G+ EN F VWMR A P F K+ + + L Y Y
Sbjct 248 MKKYPDGYTDENLPDIHTWEEFQVWMRTAAFPKFYKLTLKNESASLPKGKYQMNIELNYP 307
Query 69 VKSFGGSKSVVVTQASWLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHRR-NPRHLGDVSW 127
+ FGG+KS V+T +GGR LG+ YL+V +C L FG++F PR +GD ++
Sbjct 308 ISLFGGTKSFVLTTNGAIGGRNMSLGVLYLIVAGLCAL-FGIIFLVKLIFQPRAMGDHTY 366
Query 128 L 128
L
Sbjct 367 L 367
> YNR048w
Length=393
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 48/102 (47%), Gaps = 0/102 (0%)
Query 28 SHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSFGGSKSVVVTQASWLG 87
F +WMR A LP F K+ + L Y+ Y V+SF G+KS V+T S +G
Sbjct 270 EQFKIWMRTAALPNFYKLAMKNETNGLGKGIYIADIELNYPVRSFYGTKSFVLTTNSIIG 329
Query 88 GRTSFLGIAYLVVGSVCLLFFGVLFYYHRRNPRHLGDVSWLR 129
LGI YL+V + LF + PR + D S+L
Sbjct 330 AGNEALGIVYLIVAGIATLFAILFLIKVIFKPRPMHDHSYLN 371
> YNL323w
Length=414
Score = 44.7 bits (104), Expect = 5e-05, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 67 YDVKSFGGSKSVVVTQASWLGGRTSFLGIAYLVVGSVCLLFFGVLFYYHRRNPRHLGDVS 126
+ V F G K + +T S LGGR FLGI YL+ G +C +L + R + D S
Sbjct 348 WPVLEFNGKKGIYLTHGSHLGGRNPFLGIVYLIGGCICAAMALILLTFWLFGGRKIADAS 407
Query 127 WL 128
L
Sbjct 408 SL 409
> Hs22042456
Length=314
Score = 34.7 bits (78), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 24 GVENSHFIVWMREAPLPVFRKVYARF 49
G N FIVWMR A P F+K+Y R
Sbjct 242 GFLNDDFIVWMRAAAFPTFKKLYGRL 267
> ECU06g1450
Length=270
Score = 33.1 bits (74), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 35/85 (41%), Gaps = 14/85 (16%)
Query 27 NSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSFG----GSKSVVVTQ 82
N F W+ AP P FRK++ T TY + G K V + Q
Sbjct 189 NERFTNWIYIAPFPSFRKLWGVIDV----------ATEGTYTLNITSRFPYGDKRVSLIQ 238
Query 83 ASWLGGRTSFLGIAYLVVGSVCLLF 107
+S +G + FL I ++VG +L
Sbjct 239 SSVIGSKNYFLSIGLILVGLSMILL 263
> CE17878
Length=2030
Score = 31.2 bits (69), Expect = 0.64, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 24/52 (46%), Gaps = 2/52 (3%)
Query 24 GVENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSFGGS 75
+ N HF + E PL + R+ AQ L LP ++ + D K+ GS
Sbjct 75 AIRNHHFAII--EGPLGCGKSFLGRYAAQTLGLPLHIMQMGDQIDSKTLFGS 124
> Hs4503373
Length=1025
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 6/48 (12%)
Query 40 PVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSF------GGSKSVVVT 81
P + R+V Q +++PF+ K+T N D+ S GG+ V T
Sbjct 688 PELVRNICRWVRQAVQIPFFAKLTPNVTDIVSIARAAKEGGANGVTAT 735
> Hs22042634
Length=403
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 30/62 (48%), Gaps = 3/62 (4%)
Query 2 DFWLFNQTYASALHMDKPGVGWGVENSHFIVWMREAPLPVFRKVYARFVAQPLKLPFYLK 61
D +FN Y + L + + G G GV N HF+ E P + K + P ++P L+
Sbjct 291 DMVIFNTNYLAQLIL-RGGTGRGVNNRHFV--GEECPPTLEIKPSTKEALTPREIPSALE 347
Query 62 VT 63
V+
Sbjct 348 VS 349
> Hs16418409
Length=189
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 91 SFLGIAYLVVGSVCLLFFGVLFYYHRRNPR 120
SF+G L +G + +L+ G YY RR R
Sbjct 149 SFVGGIVLTLGVLSILYIGCKMYYSRRGIR 178
> At1g08350
Length=589
Score = 27.7 bits (60), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 4/48 (8%)
Query 91 SFLGIAYLVVGSVCLLFFGVLFYYH----RRNPRHLGDVSWLRKALYS 134
+ L I +L++G V FG+L + +RNPR + +W R+ LY
Sbjct 403 TLLNIPFLMLGGVLGNRFGLLEFQPPSAVKRNPREIPPQNWYRRKLYQ 450
> CE02496
Length=1084
Score = 27.7 bits (60), Expect = 6.8, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 6/48 (12%)
Query 40 PVFRKVYARFVAQPLKLPFYLKVTNNTYDVKSF------GGSKSVVVT 81
P K R+V +K+PF+ K+T N DV+ GG+ V T
Sbjct 727 PEIVKEICRWVRACVKIPFFPKMTPNITDVREIARAARDGGASGVTAT 774
> CE28398
Length=728
Score = 27.3 bits (59), Expect = 8.0, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query 69 VKSFGGSKSVVVTQASWLGGRTSFLGIAYLVVGSVCLLF 107
+ S+ GSK ++ W+GG + I YL++G+ LF
Sbjct 60 IMSYFGSKG---NRSRWIGGGVLLMSITYLMLGTPNFLF 95
Lambda K H
0.328 0.142 0.470
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1393086308
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40