bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2822_orf1
Length=111
Score E
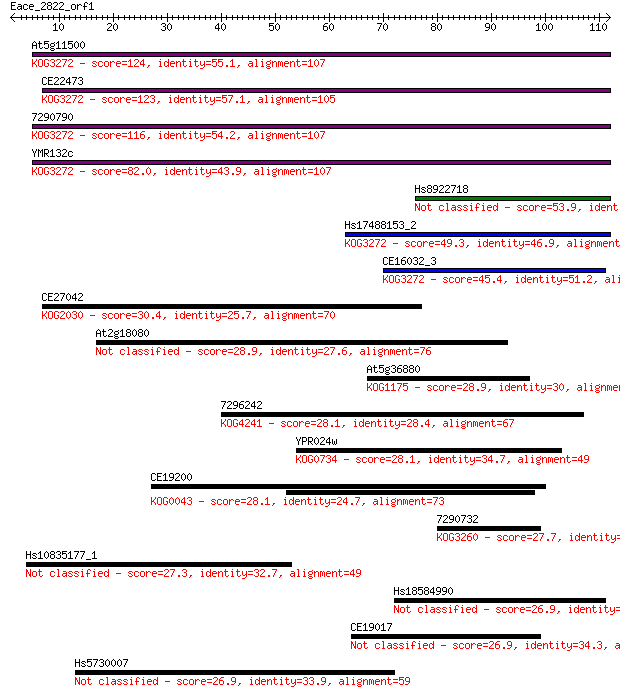
Sequences producing significant alignments: (Bits) Value
At5g11500 124 5e-29
CE22473 123 8e-29
7290790 116 9e-27
YMR132c 82.0 2e-16
Hs8922718 53.9 7e-08
Hs17488153_2 49.3 2e-06
CE16032_3 45.4 3e-05
CE27042 30.4 0.95
At2g18080 28.9 2.4
At5g36880 28.9 2.7
7296242 28.1 3.7
YPR024w 28.1 4.0
CE19200 28.1 4.6
7290732 27.7 5.8
Hs10835177_1 27.3 6.9
Hs18584990 26.9 8.6
CE19017 26.9 8.8
Hs5730007 26.9 9.0
> At5g11500
Length=215
Score = 124 bits (310), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 59/109 (54%), Positives = 79/109 (72%), Gaps = 3/109 (2%)
Query 5 MVFTYQCSTPG--YEIYMGRDKYENEDLIVHAFPEDVWFHVSELSSAHVYVRMPFGQTDY 62
MVF ++ Y I+MG DK+ENE+LI + FPEDVWFHV ++SSAHVY+R+ GQ+ +
Sbjct 1 MVFYFKARPDAGDYTIFMGLDKFENEELIKYGFPEDVWFHVDKMSSAHVYLRLHRGQS-F 59
Query 63 ESLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQV 111
+ + V+E+ QL K NSI+G K + V +VYTP SNLKKTASM+ GQV
Sbjct 60 DDISEGVLEDCAQLVKANSIQGNKVNNVDVVYTPWSNLKKTASMDVGQV 108
> CE22473
Length=210
Score = 123 bits (309), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 60/105 (57%), Positives = 73/105 (69%), Gaps = 1/105 (0%)
Query 7 FTYQCSTPGYEIYMGRDKYENEDLIVHAFPEDVWFHVSELSSAHVYVRMPFGQTDYESLP 66
F+ + P IYMG DK ENEDLI + +PEDVWFHV +LSSAHVY+R+ G T +S+P
Sbjct 5 FSSNTTNPPTMIYMGVDKVENEDLIKYGWPEDVWFHVDKLSSAHVYLRLHSGMT-IDSIP 63
Query 67 AEVIEEACQLTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQV 111
++ + CQL K NSIEGCK + V IVYT SNLKKT M GQV
Sbjct 64 EALLIDCCQLVKQNSIEGCKLNNVAIVYTMWSNLKKTGDMAVGQV 108
> 7290790
Length=209
Score = 116 bits (291), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 58/109 (53%), Positives = 77/109 (70%), Gaps = 3/109 (2%)
Query 5 MVFTYQCST--PGYEIYMGRDKYENEDLIVHAFPEDVWFHVSELSSAHVYVRMPFGQTDY 62
MVF ++ + P +YMGRDK+ENE+LI +PEDVWFHV +LSSAHVY+R+ GQT
Sbjct 1 MVFYFKSNVVQPPAMLYMGRDKHENEELIRWGWPEDVWFHVHDLSSAHVYLRLRKGQT-I 59
Query 63 ESLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQV 111
+ +P +V+ +A QL K NSI+G K + + +VYT NLKKT ME GQV
Sbjct 60 DDIPTDVLVDAAQLCKANSIQGNKVNNLEVVYTMWENLKKTPDMEPGQV 108
> YMR132c
Length=208
Score = 82.0 bits (201), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 47/114 (41%), Positives = 68/114 (59%), Gaps = 8/114 (7%)
Query 5 MVFTYQC-----STPGYEIYMGRDKYENEDLIVHAFPED--VWFHVSELSSAHVYVRMPF 57
MV+ Y+ STP Y+I MG+DK+EN+ LI ++ E VWFH + SS HVY+++
Sbjct 1 MVYFYESKPTEYSTP-YQIVMGKDKFENDLLIKWSYRELNYVWFHADKYSSGHVYLKLRP 59
Query 58 GQTDYESLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQV 111
+ + +P EVI + QL K SI+G K + TI+ TP NL+K M G+V
Sbjct 60 NEKTIDDIPQEVICDCLQLCKSESIQGNKMPQCTILITPWHNLRKNRYMNPGEV 113
> Hs8922718
Length=140
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 23/36 (63%), Positives = 28/36 (77%), Gaps = 0/36 (0%)
Query 76 LTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQV 111
L K NSI+GCK + V +VYTP SNLKKTA M+ GQ+
Sbjct 6 LVKANSIQGCKMNNVNVVYTPWSNLKKTADMDVGQI 41
> Hs17488153_2
Length=238
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/49 (46%), Positives = 32/49 (65%), Gaps = 1/49 (2%)
Query 63 ESLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQV 111
E +P EV+ + L K NS +GCK + V +V TP SNLKKTA ++ Q+
Sbjct 5 EDIPKEVLTDCAHLVKANSTQGCKNN-VNVVNTPRSNLKKTADVDVRQI 52
> CE16032_3
Length=148
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/41 (51%), Positives = 27/41 (65%), Gaps = 0/41 (0%)
Query 70 IEEACQLTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQ 110
+EE CQL K NSI+G K +V + YT NLKK M+TG+
Sbjct 31 LEECCQLVKKNSIQGVKMEKVEVNYTLKENLKKVKGMQTGE 71
> CE27042
Length=899
Score = 30.4 bits (67), Expect = 0.95, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 35/71 (49%), Gaps = 4/71 (5%)
Query 7 FTYQCSTPGYEIYMGRDKYENEDLIVHAF-PEDVWFHVSELSSAHVYVRMPFGQTDYESL 65
F + S+ G+ + GRD +NE L+ P D++ H ++ V +R ++ +
Sbjct 472 FRWFISSEGFIVVAGRDAQQNELLVKKYLRPNDIYMHADVRGASSVVIR---NKSFDAEI 528
Query 66 PAEVIEEACQL 76
P + + EA Q+
Sbjct 529 PPKTLTEAAQM 539
> At2g18080
Length=365
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 33/77 (42%), Gaps = 16/77 (20%)
Query 17 EIYMGRDKYENEDLIVHAFPEDVWFHVSELSSAHVYVRMPFGQTDYESLPAEVIEEACQL 76
+Y G D+ +I ED W H S+ +S H +P+ +I+ C+
Sbjct 265 NLYYGGDRLAATKIIFTNGSEDPWRHASKQNSTH-------------EMPSYIIK--CRN 309
Query 77 TKYNS-IEGCKQSEVTI 92
+ S I GC QS + I
Sbjct 310 CGHGSDIRGCPQSPMVI 326
> At5g36880
Length=693
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 9/30 (30%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 67 AEVIEEACQLTKYNSIEGCKQSEVTIVYTP 96
+E+++ CQL Y G K+ + ++Y P
Sbjct 151 SELLQRVCQLANYLKDNGVKKGDAVVIYLP 180
> 7296242
Length=248
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 43/70 (61%), Gaps = 5/70 (7%)
Query 40 WFHVSELSSAHVY-VRMPFGQTDY--ESLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTP 96
+FH+S +++ ++ VR+ + + +S +++IE+A + T+Y +I S IV++P
Sbjct 96 FFHLSSITADDIFRVRVQLHKQNLHLKSYGSKIIEQAVKNTRYEAIVPLFHSNHCIVFSP 155
Query 97 ASNLKKTASM 106
+ +KTA++
Sbjct 156 --DPEKTAAL 163
> YPR024w
Length=747
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 3/49 (6%)
Query 54 RMPFGQTDYESLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTPASNLKK 102
R+ G +YE+L A IE+ C+ K + + K S T+V P S+ +K
Sbjct 687 RLAQGLIEYETLDAHEIEQVCKGEKLDKL---KTSTNTVVEGPDSDERK 732
> CE19200
Length=478
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 18/73 (24%), Positives = 30/73 (41%), Gaps = 5/73 (6%)
Query 27 NEDLIVHAFPEDVWFHVSELSSAHVYVRMPFGQTDYESLPAEVIEEACQLTKYNSIEGCK 86
NED PED W V + ++P Q ++ + P ++ C L N GC+
Sbjct 216 NED---EELPEDPWL-VRRRGAPRTAPKLP-DQREFINRPPMLVYRCCWLDHMNDFHGCR 270
Query 87 QSEVTIVYTPASN 99
+ +Y S+
Sbjct 271 MKRIFKMYVYCSD 283
Score = 26.9 bits (58), Expect = 9.4, Method: Composition-based stats.
Identities = 11/46 (23%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 52 YVRMPFGQTDYESLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTPA 97
+ R FG+TD+ + ++++EE T++ + + T+V P+
Sbjct 350 FTRQQFGETDFANYSSQILEEGPPTTQFFNSPSILKFRCTMVDAPS 395
> 7290732
Length=230
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 9/19 (47%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 80 NSIEGCKQSEVTIVYTPAS 98
N ++GC + VT+ YTP S
Sbjct 87 NGVQGCTEENVTVTYTPNS 105
> Hs10835177_1
Length=541
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 26/53 (49%), Gaps = 4/53 (7%)
Query 4 KMVFTYQCSTPGYEIYMGRDKYENEDLIVHAFPED----VWFHVSELSSAHVY 52
+M T C T G EIY+ DK + +D+ + + E+ VW + S V+
Sbjct 254 RMDRTAGCVTGGEEIYLLCDKVQKDDIQIRFYEEEENGGVWEGFGDFSPTDVH 306
> Hs18584990
Length=443
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 72 EACQLTKYNSIEGCKQSEVTIVYTPASNLKKTASMETGQ 110
E CQL K++S+E C E+ + SNL +T + Q
Sbjct 29 EGCQLIKFDSVEVCVCCELQHQSSGCSNLGETLKLNPLQ 67
> CE19017
Length=362
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 64 SLPAEVIEEACQLTKYNSIEGCKQSEVTIVYTPAS 98
+L + IEE K++ + GCK+SE++++ T S
Sbjct 312 NLKLKSIEEKNYAEKFSELLGCKKSEMSVIPTEMS 346
> Hs5730007
Length=579
Score = 26.9 bits (58), Expect = 9.0, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 31/61 (50%), Gaps = 5/61 (8%)
Query 13 TPGYEIYMGRDKYENEDLIVHAFPEDVWFHVSELSSAHVY--VRMPFGQTDYESLPAEVI 70
T G E+Y+ DK + ED+ V F W ++ S A V+ + + F YE L E++
Sbjct 316 TGGEELYLLCDKVQKEDISV-VFSRASWEGRADFSQADVHRQIAIVFKTPPYEDL--EIV 372
Query 71 E 71
E
Sbjct 373 E 373
Lambda K H
0.315 0.130 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40