bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2834_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
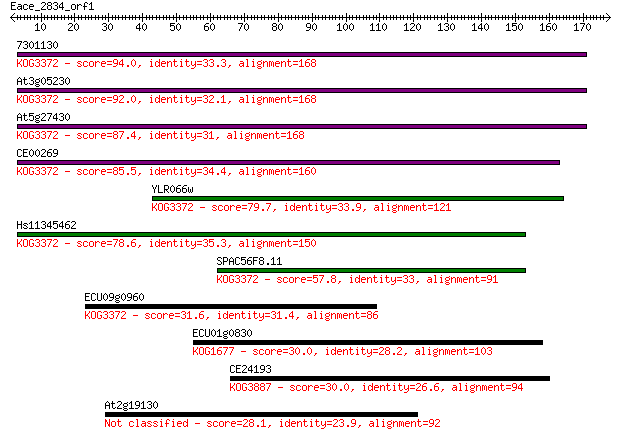
7301130 94.0 1e-19
At3g05230 92.0 5e-19
At5g27430 87.4 1e-17
CE00269 85.5 5e-17
YLR066w 79.7 2e-15
Hs11345462 78.6 6e-15
SPAC56F8.11 57.8 1e-08
ECU09g0960 31.6 0.91
ECU01g0830 30.0 2.3
CE24193 30.0 2.6
At2g19130 28.1 9.9
> 7301130
Length=179
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 56/175 (32%), Positives = 87/175 (49%), Gaps = 8/175 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFFLVGPISGSVSA-----REVYGFGYNYAL 57
M L R NA + L L S+ FL +++ + V +G +
Sbjct 1 MHTVLTRGNATVAYTLSVLACLTFSCFLSTVFLDYRTDANINTVRVLVKNVPDYGASRE- 59
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
D ++FD++ +L G+F WN KQLF+++ AEY++P + NQVV++D+II AVLD
Sbjct 60 KHDLGFVTFDLQTNLTGIFNWNVKQLFLYLTAEYQTPANQLNQVVLWDKIILRGDNAVLD 119
Query 118 LANVPAKYHLRDKGKGLR-GREITLKLQVVYHPIVGRIYT-QTVASSTFRMPAQY 170
N+ KY+ D G GL+ R ++L L P G + + Q F+ PA Y
Sbjct 120 FKNMNTKYYFWDDGNGLKDNRNVSLYLSWNIIPNAGLLPSVQATGKHLFKFPADY 174
> At3g05230
Length=167
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 54/169 (31%), Positives = 86/169 (50%), Gaps = 4/169 (2%)
Query 3 MENYLNRANAVFCSLMVSLG-VLALGNIASSFFLVGPISGSVSAREVYGFGYNYALSGDQ 61
M + RANA+ + +L + A+ + + F P S + + F + D+
Sbjct 1 MHTFGYRANALLTFAVTALAFICAIASFSDKFSNQNP-SAEIQILNINRFK-KQSHGNDE 58
Query 62 AVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLANV 121
L+ DI ADL+ LF WN KQ+FVFV AEYE+P++ NQV ++D II + A + V
Sbjct 59 VSLTLDISADLQSLFTWNTKQVFVFVAAEYETPKNSLNQVSLWDAIIPAKEHAKFRI-QV 117
Query 122 PAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTVASSTFRMPAQY 170
KY D+G+ LRG++ L L P G+++ + + +P Y
Sbjct 118 SNKYRFIDQGQNLRGKDFNLTLHWHVMPKTGKMFADKIVLPGYSLPDAY 166
> At5g27430
Length=167
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 52/169 (30%), Positives = 85/169 (50%), Gaps = 4/169 (2%)
Query 3 MENYLNRANAVFCSLMVSLG-VLALGNIASSFFLVGPISGSVSAREVYGFGYNYALSGDQ 61
M ++ RANA+ + L + A+ + + +F P S + + F D+
Sbjct 1 MHSFGYRANALLTFAVTILAFICAIASFSDNFSNQNP-SAQIQILNINWFQ-KQPHGNDE 58
Query 62 AVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLDLANV 121
L+ +I ADL+ LF WN KQ+F FV AEYE+ ++ NQV ++D II + A + +
Sbjct 59 VSLTLNITADLQSLFTWNTKQVFAFVAAEYETSKNALNQVSLWDAIIPEKEHAKFWI-QI 117
Query 122 PAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTVASSTFRMPAQY 170
KY D+G LRG++ L L P G+++ + S +R+P Y
Sbjct 118 SNKYRFIDQGHNLRGKDFNLTLHWHVMPKTGKMFADKIVMSGYRLPNAY 166
> CE00269
Length=180
Score = 85.5 bits (210), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 55/166 (33%), Positives = 83/166 (50%), Gaps = 9/166 (5%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFFLVGPIS-----GSVSAREVYGFGYNYAL 57
M N L+RANA+ + + + S+ FL + V R V + +
Sbjct 1 MHNLLSRANALLAFTLWVMAAVTAACFLSTVFLDYTVPTKLTVNDVKVRNVVDYATDEQ- 59
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
D A L+F++K D +F WN KQLFV++VAEY+S + NQVV++DRI+ V+D
Sbjct 60 QADLATLNFNLKVDFSKIFNWNVKQLFVYLVAEYKSKVNEVNQVVLWDRIVERADRVVMD 119
Query 118 LANVPAKYHLRDKGKG-LRGREITLKLQVVYHPIVGRIYTQTVASS 162
V +KY+ D G L + +T L+ Y+ I Y + V SS
Sbjct 120 EIGVKSKYYFLDDGTNLLNHKNVTFVLR--YNVIPNSGYLRLVQSS 163
> YLR066w
Length=184
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 41/123 (33%), Positives = 70/123 (56%), Gaps = 2/123 (1%)
Query 43 VSAREVYGFGYNYALSGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVV 102
++ R FG + + + FD+ DL LF WN KQ+FV++ AEY S + T++V
Sbjct 53 INVRTSRYFGSQRGKAKENMKIKFDLNTDLTPLFNWNTKQVFVYLTAEYNSTEKITSEVT 112
Query 103 VFDRIITNESEAVLDLANVPAKYHLRDKGKG-LRGREITLKLQVVYHPIVGRI-YTQTVA 160
+D+II ++ +AV+D+ ++ +KY + D G G+++ KL P VG + Y +TV
Sbjct 113 FWDKIIKSKDDAVIDVNDLRSKYSIWDIEDGKFEGKDLVFKLHWNVQPWVGLLTYGETVG 172
Query 161 SST 163
+ T
Sbjct 173 NYT 175
> Hs11345462
Length=180
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 53/156 (33%), Positives = 78/156 (50%), Gaps = 7/156 (4%)
Query 3 MENYLNRANAVFCSLMVSLGVLALGNIASSFF--LVGPISGSVS---AREVYGFGYNYAL 57
M L+RAN++F + + L G ++ F P+ VS + V F
Sbjct 1 MNTVLSRANSLFAFSLSVMAALTFGCFITTAFKDRSVPVRLHVSRIMLKNVEDFTGPRER 60
Query 58 SGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVLD 117
S D ++FDI ADL +F WN KQLF+++ AEY + + NQVV++D+I+ L
Sbjct 61 S-DLGFITFDITADLENIFDWNVKQLFLYLSAEYSTKNNALNQVVLWDKIVLRGDNPKLL 119
Query 118 LANVPAKYHLRDKGKGLRG-REITLKLQVVYHPIVG 152
L ++ KY D G GL+G R +TL L P G
Sbjct 120 LKDMKTKYFFFDDGNGLKGNRNVTLTLSWNVVPNAG 155
> SPAC56F8.11
Length=185
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 52/95 (54%), Gaps = 5/95 (5%)
Query 62 AVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEAVL----D 117
A + F++ ADL L+ WN K + V++VA Y + +H NQVVV+D+I+++ E+ +
Sbjct 69 AQVKFNMDADLSELWDWNTKHVVVYLVASYSTEKHEKNQVVVWDKILSSPEESKMFMKDT 128
Query 118 LANVPAKYHLRDKGKGLRGREITLKLQVVYHPIVG 152
L+N+ A + + G+ T L P +G
Sbjct 129 LSNIQA-HPFNEYSNQFEGKNATYTLHWTVSPKMG 162
> ECU09g0960
Length=151
Score = 31.6 bits (70), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 39/87 (44%), Gaps = 10/87 (11%)
Query 23 VLALGNIASSFFLV-GPISGSVSAREVYGFGYNYALSGDQAVLSFDIKADLRGLFQWNAK 81
+L L SFF+ P + S+ RE Y G V+ F+ DL F +N K
Sbjct 21 ILLLAVFLFSFFVTKAPPASSLRIRE-------YKAYGKTQVVLFEPSIDLSSQFHFNLK 73
Query 82 QLFVFVVAEYESPQHPTNQVVVFDRII 108
Q+FV+V Y + V++ RII
Sbjct 74 QIFVYVRVLYGGRSDRSE--VIWSRII 98
> ECU01g0830
Length=346
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 41/103 (39%), Gaps = 10/103 (9%)
Query 55 YALSGDQAVLSFDIKADLRGLFQWNAKQLFVFVVAEYESPQHPTNQVVVFDRIITNESEA 114
Y GD+ + KA+LR Q + K +E P + F I
Sbjct 126 YCRYGDKCQFAHS-KAELR-YVQRHPKYKTETCKTFWEEGSCPYGKRCCFIHIPNT---- 179
Query 115 VLDLANVPAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQ 157
D+AN+P RD GK G TL +++ HP IYT+
Sbjct 180 --DIANLPIHGRQRDSGKNCTG--YTLGIEIDEHPHDELIYTK 218
> CE24193
Length=338
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 25/105 (23%), Positives = 47/105 (44%), Gaps = 14/105 (13%)
Query 66 FDIKADLRGLFQWNAKQLFVF-----------VVAEYESPQHPTNQVVVFDRIITNESEA 114
FD D G+FQ LF+ + EY + NQ + F+ + ++++
Sbjct 88 FDDSLDPVGVFQKCEALLFIIDAQAELQEPIATLVEYFCRAYKINQNIKFE-VFVHKADG 146
Query 115 VLDLANVPAKYHLRDKGKGLRGREITLKLQVVYHPIVGRIYTQTV 159
+ + A V K+++ + K +I + LQV YH + IY ++
Sbjct 147 LTEEARVETKFNIYHQVKETIKDQIDVDLQVTYH--LTSIYDHSI 189
> At2g19130
Length=828
Score = 28.1 bits (61), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 46/92 (50%), Gaps = 5/92 (5%)
Query 29 IASSFFLVGPISGSVSAREVYGFGYNYALSGDQAVLSFDIKADLRGLFQWNAKQLFVFVV 88
+ S FF+ I GS + + G ++ LSGDQ ++S D ++ G F+ + F ++
Sbjct 7 LTSFFFICFFIHGSSAVDTISG---DFTLSGDQTIVSSDGTYEM-GFFKPGSSSNF-YIG 61
Query 89 AEYESPQHPTNQVVVFDRIITNESEAVLDLAN 120
Y+ V D+ +++++ +V ++N
Sbjct 62 MWYKQLSQTILWVANRDKAVSDKNSSVFKISN 93
Lambda K H
0.322 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2743263016
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40