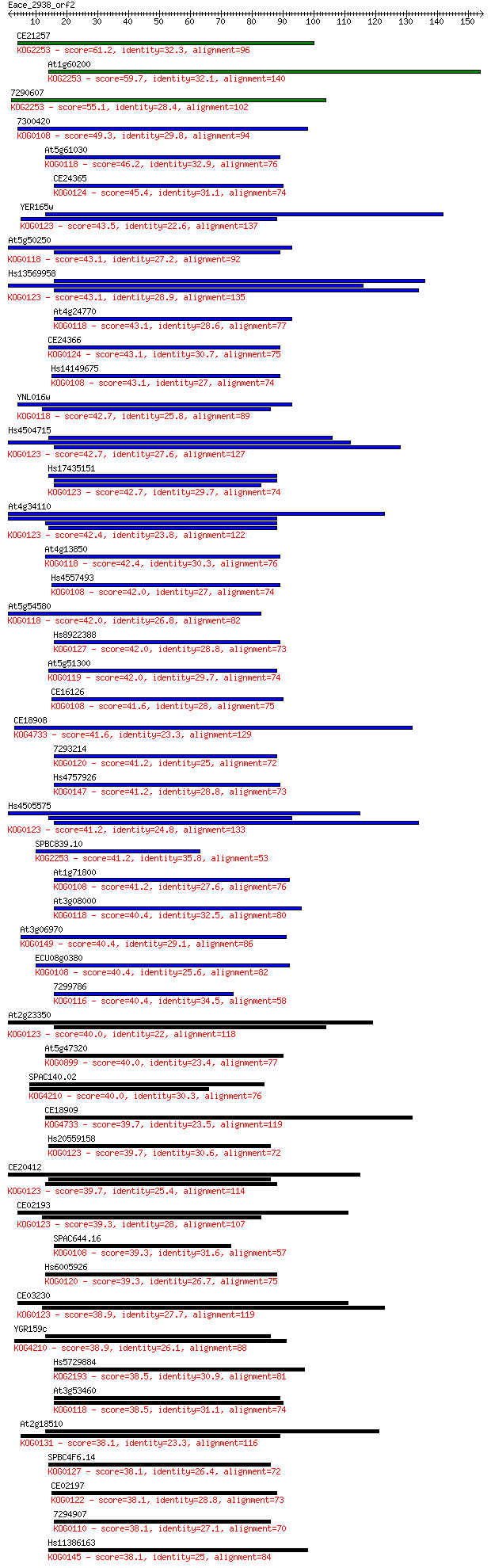
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2938_orf2
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
CE21257 61.2 7e-10
At1g60200 59.7 2e-09
7290607 55.1 5e-08
7300420 49.3 3e-06
At5g61030 46.2 2e-05
CE24365 45.4 5e-05
YER165w 43.5 1e-04
At5g50250 43.1 2e-04
Hs13569958 43.1 2e-04
At4g24770 43.1 2e-04
CE24366 43.1 2e-04
Hs14149675 43.1 2e-04
YNL016w 42.7 3e-04
Hs4504715 42.7 3e-04
Hs17435151 42.7 3e-04
At4g34110 42.4 4e-04
At4g13850 42.4 4e-04
Hs4557493 42.0 4e-04
At5g54580 42.0 5e-04
Hs8922388 42.0 5e-04
At5g51300 42.0 5e-04
CE16126 41.6 6e-04
CE18908 41.6 6e-04
7293214 41.2 7e-04
Hs4757926 41.2 8e-04
Hs4505575 41.2 9e-04
SPBC839.10 41.2 9e-04
At1g71800 41.2 9e-04
At3g08000 40.4 0.001
At3g06970 40.4 0.001
ECU08g0380 40.4 0.001
7299786 40.4 0.001
At2g23350 40.0 0.002
At5g47320 40.0 0.002
SPAC140.02 40.0 0.002
CE18909 39.7 0.002
Hs20559158 39.7 0.002
CE20412 39.7 0.003
CE02193 39.3 0.003
SPAC644.16 39.3 0.003
Hs6005926 39.3 0.003
CE03230 38.9 0.004
YGR159c 38.9 0.004
Hs5729884 38.5 0.005
At3g53460 38.5 0.006
At2g18510 38.1 0.006
SPBC4F6.14 38.1 0.007
CE02197 38.1 0.007
7294907 38.1 0.007
Hs11386163 38.1 0.008
> CE21257
Length=712
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 50/96 (52%), Gaps = 1/96 (1%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
QQ QQ+ ++ +++GNI+ ++F+R +L E G V W R +G+L FGFC F
Sbjct 37 QQSSQQKPDSSPVFVGNISDKCSDEFIRKILNECGNVASWKRIKGS-NGKLQGFGFCHFT 95
Query 64 EPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAK 99
+ + + L L + L V +EKVR + K
Sbjct 96 DLEGTLRALRILHEFHLGDKKLTVKPDEKVRDDLRK 131
> At1g60200
Length=781
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 45/152 (29%), Positives = 75/152 (49%), Gaps = 13/152 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
TT+YIG I +NDFM ++L G V R DP + + FGF +F + +
Sbjct 55 TTIYIGKIAT-VENDFMMSILEFCGHVKSCLRAEDPTTKKPKGFGFYEFESAEGILRAIR 113
Query 74 CLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNK-TREQVEKEIEEEYKRL- 131
L R + G+ L+VN N+ + + K E++I +A +++ T+E + E E +L
Sbjct 114 LLTQRTIDGQELLVNVNQATKEYLLKYVEKKIETAKKAKESQGTKENQAEGPESEQDKLE 173
Query 132 -------KDGVQQI---LDKKNASLPEDDETE 153
KDG +I +D N+++ D+E E
Sbjct 174 SADNETGKDGESKIKENIDIANSAVLTDEERE 205
> 7290607
Length=998
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/104 (27%), Positives = 52/104 (50%), Gaps = 10/104 (9%)
Query 2 QQQQQQQQQQRAT--TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGF 59
Q+ + Q Q R T+++GNI++ ++ +L G V+ W R ++ FGF
Sbjct 47 QRPPEPQPQFRGPIITVFVGNISERVPEALLKRILNACGVVINWKR--------VSTFGF 98
Query 60 CDFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEE 103
C+F P+AA + V L+ ++ G+ LV + K + I E+
Sbjct 99 CEFDGPIAAMRAVRLLSEMEIDGKKLVAKVDAKNKVLIEDYKEQ 142
> 7300420
Length=399
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 49/96 (51%), Gaps = 2/96 (2%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
Q+Q + ++++GNI A + ++ + E G VL D SG+ FGFC++
Sbjct 6 QEQSIMDKSMRSVFVGNIPYEATEEKLKEIFSEVGPVLSLKLVFDRESGKPKGFGFCEYK 65
Query 64 EPLAAAKCVACLAGRKLHGRSLVVN--CNEKVRAKI 97
+ A + L G ++ GR+L V+ C EK R ++
Sbjct 66 DQETALSAMRNLNGYEIGGRTLRVDNACTEKSRMEM 101
> At5g61030
Length=309
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 38/76 (50%), Gaps = 0/76 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
++ L+IG + D D +R ++G V+ D +G+ FGF F AA+ +
Sbjct 39 SSKLFIGGMAYSMDEDSLREAFTKYGEVVDTRVILDRETGRSRGFGFVTFTSSEAASSAI 98
Query 73 ACLAGRKLHGRSLVVN 88
L GR LHGR + VN
Sbjct 99 QALDGRDLHGRVVKVN 114
> CE24365
Length=339
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 0/74 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+Y+G+I+ D +R FG + N DP +G F F ++ P AA +
Sbjct 104 IYVGSISFEIREDMLRRAFDPFGPIKSINMSWDPATGHHKTFAFVEYEVPEAALLAQESM 163
Query 76 AGRKLHGRSLVVNC 89
G+ L GR+L VN
Sbjct 164 NGQMLGGRNLKVNS 177
> YER165w
Length=577
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 59/129 (45%), Gaps = 1/129 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+ ++I N+ DN + + FG +L +D +G+ FGF F E AA + +
Sbjct 125 SGNIFIKNLHPDIDNKALYDTFSVFGDILSSKIATDE-NGKSKGFGFVHFEEEGAAKEAI 183
Query 73 ACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRLK 132
L G L+G+ + V + + + +++ E + T KN E +++ +E + +
Sbjct 184 DALNGMLLNGQEIYVAPHLSRKERDSQLEETKAHYTNLYVKNINSETTDEQFQELFAKFG 243
Query 133 DGVQQILDK 141
V L+K
Sbjct 244 PIVSASLEK 252
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 41/83 (49%), Gaps = 1/83 (1%)
Query 5 QQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAE 64
Q ++ + T LY+ NI ++ + L +FG ++ + + D G+L FGF ++ +
Sbjct 210 QLEETKAHYTNLYVKNINSETTDEQFQELFAKFGPIVSASLEKD-ADGKLKGFGFVNYEK 268
Query 65 PLAAAKCVACLAGRKLHGRSLVV 87
A K V L +L+G L V
Sbjct 269 HEDAVKAVEALNDSELNGEKLYV 291
> At5g50250
Length=289
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 44/92 (47%), Gaps = 0/92 (0%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+ ++Q + A +Y+GN+ D+ + L E G+V+ SD +G+ FGF
Sbjct 194 SRPERQPRVYDAAFRIYVGNLPWDVDSGRLERLFSEHGKVVDARVVSDRETGRSRGFGFV 253
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEK 92
+ +A L G+ L GR++ VN E+
Sbjct 254 QMSNENEVNVAIAALDGQNLEGRAIKVNVAEE 285
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 35/77 (45%), Gaps = 8/77 (10%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRV----LRWNRQSDPISGQLAAFGFCDFAEPLAAAKC 71
L++GN+ D+ + L + G V + +NR +D Q FGF + A K
Sbjct 115 LFVGNLPYDVDSQALAMLFEQAGTVEISEVIYNRDTD----QSRGFGFVTMSTVEEAEKA 170
Query 72 VACLAGRKLHGRSLVVN 88
V +++GR L VN
Sbjct 171 VEKFNSFEVNGRRLTVN 187
> Hs13569958
Length=631
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 61/121 (50%), Gaps = 9/121 (7%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+YI N + D++ +++L G+FG L +D SG+ FGF F A K V +
Sbjct 193 VYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVDEM 251
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTE-ERILATMRAYKNKTREQVEKEIEEEYKRLKDG 134
G++L+G+ + V +K K+ + TE +R M+ + TR QV + K L DG
Sbjct 252 NGKELNGKQIYVGRAQK---KVERQTELKRTFEQMKQDR-ITRYQV---VNLYVKNLDDG 304
Query 135 V 135
+
Sbjct 305 I 305
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 55/117 (47%), Gaps = 4/117 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + + + LY+ N+ D++ +R FG + + G+ FGF
Sbjct 281 EQMKQDRITRYQVVNLYVKNLDDGIDDERLRKAFSPFGTITSAKVMME--GGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKNK 115
F+ P A K V + GR + + L V ++ + A +T E + +A++RA N+
Sbjct 339 CFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEERQAYLTNEYMQRMASVRAVPNQ 395
Score = 32.3 bits (72), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 28/118 (23%), Positives = 54/118 (45%), Gaps = 9/118 (7%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+++ N+ K +N + + + FG +L N D + +GF F AA + + +
Sbjct 101 IFVKNLDKSINNKALYDTVSAFGNILSCNVVCDENGSK--GYGFVHFETHEAAERAIKKM 158
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRLKD 133
G L+GR + V + K K E + A + + N + +++++E RLKD
Sbjct 159 NGMLLNGRKVFVG-----QFKSRKEREAELGARAKEFPNVYIKNFGEDMDDE--RLKD 209
> At4g24770
Length=329
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 0/77 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+Y+GN+ DN + L E G+V+ D +G+ FGF ++ + ++ L
Sbjct 246 VYVGNLPWDVDNGRLEQLFSEHGKVVEARVVYDRETGRSRGFGFVTMSDVDELNEAISAL 305
Query 76 AGRKLHGRSLVVNCNEK 92
G+ L GR++ VN E+
Sbjct 306 DGQNLEGRAIRVNVAEE 322
> CE24366
Length=749
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 36/75 (48%), Gaps = 0/75 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
+ +Y+G+I+ D +R FG + N DP +G F F ++ P AA
Sbjct 102 SRIYVGSISFEIREDMLRRAFDPFGPIKSINMSWDPATGHHKTFAFVEYEVPEAALLAQE 161
Query 74 CLAGRKLHGRSLVVN 88
+ G+ L GR+L VN
Sbjct 162 SMNGQMLGGRNLKVN 176
> Hs14149675
Length=616
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 17 SVFVGNIPYEATEEQLKDIFSEVGSVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 76
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 77 LNGREFSGRALRVD 90
> YNL016w
Length=453
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 40/89 (44%), Gaps = 0/89 (0%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
Q QQ L++G++ + D++ +RN +F L + D +G +GF F
Sbjct 152 QSQQSSSDDTFNLFVGDLNVNVDDETLRNAFKDFPSYLSGHVMWDMQTGSSRGYGFVSFT 211
Query 64 EPLAAAKCVACLAGRKLHGRSLVVNCNEK 92
A + + G+ L+GR L +N K
Sbjct 212 SQDDAQNAMDSMQGQDLNGRPLRINWAAK 240
Score = 31.6 bits (70), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 29/74 (39%), Gaps = 6/74 (8%)
Query 12 RATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKC 71
R TT YIGNI A + L FG +L + + F + AA C
Sbjct 339 RVTTAYIGNIPHFATEADLIPLFQNFGFILDFKHYPE------KGCCFIKYDTHEQAAVC 392
Query 72 VACLAGRKLHGRSL 85
+ LA GR+L
Sbjct 393 IVALANFPFQGRNL 406
> Hs4504715
Length=644
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 50/99 (50%), Gaps = 8/99 (8%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ ++ L +FG+ L DP +G+ FGF + + A K V
Sbjct 191 TNVYIKNFGEEVDDESLKELFSQFGKTLSVKVMRDP-NGKSKGFGFVSYEKHEDANKAVE 249
Query 74 CLAGRKLHGRSLVVNCNEK-------VRAKIAKVTEERI 105
+ G+++ G+ + V +K ++ K ++ +ERI
Sbjct 250 EMNGKEISGKIIFVGRAQKKVERQAELKRKFEQLKQERI 288
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 53/113 (46%), Gaps = 4/113 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q++ + + LYI N+ D++ +R FG + + G+ FGF
Sbjct 281 EQLKQERISRYQGVNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--DGRSKGFGFV 338
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRA 111
F+ P A K V + GR + + L V ++ + A +T + + +A MRA
Sbjct 339 CFSSPEEATKAVTEMNGRIVGSKPLYVALAQRKEERKAHLTNQYMQRVAGMRA 391
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 26/112 (23%), Positives = 46/112 (41%), Gaps = 7/112 (6%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + + FG +L D + + F F AA K + +
Sbjct 101 VFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSK--GYAFVHFETQEAADKAIEKM 158
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEE 127
G L+ R + V R K K E + A + + N + +E+++E
Sbjct 159 NGMLLNDRKVFVG-----RFKSRKEREAELGAKAKEFTNVYIKNFGEEVDDE 205
> Hs17435151
Length=487
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 39/74 (52%), Gaps = 1/74 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N D++ ++++ ++G+ L +D SG+ FGF F AA K V
Sbjct 307 TNVYIKNFGGDMDDERLKDVFSKYGKTLSVKVMTDS-SGKSKGFGFVSFDSHEAAKKAVE 365
Query 74 CLAGRKLHGRSLVV 87
+ GR ++G+ + V
Sbjct 366 EMNGRDINGQLIFV 379
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
LYI N+ D++ +RN FG + R + GQ FG F+ P A K + +
Sbjct 412 LYIKNLDDTIDDEKLRNEFSSFGSISRVKVMQEE--GQSKGFGLICFSSPEDATKAMTEM 469
Query 76 AGRKLHGRSLVV 87
GR L + L +
Sbjct 470 NGRILGSKPLSI 481
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 29/67 (43%), Gaps = 2/67 (2%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + FG++L SD + + F F AA + + +
Sbjct 217 VFIKNLDKSIDNKTLYEHFSAFGKILSSKVMSDDQGSK--GYAFVHFQNQSAADRAIEEM 274
Query 76 AGRKLHG 82
G+ L G
Sbjct 275 NGKLLKG 281
> At4g34110
Length=629
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 29/122 (23%), Positives = 55/122 (45%), Gaps = 9/122 (7%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q++ + + T +Y+ N+ + +D ++N GE+G++ D G+ FGF
Sbjct 202 RQERDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDG-EGKSKGFGFV 260
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQV 120
+F AA+ V L G K + V + + K + TE R+ Y+ +E
Sbjct 261 NFENADDAARAVESLNGHKFDDKEWYVG---RAQKKSERETELRV-----RYEQNLKEAA 312
Query 121 EK 122
+K
Sbjct 313 DK 314
Score = 35.8 bits (81), Expect = 0.030, Method: Composition-based stats.
Identities = 20/87 (22%), Positives = 42/87 (48%), Gaps = 1/87 (1%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q ++ + +++ LY+ N+ ++ ++ + FG V DP +G GF
Sbjct 305 EQNLKEAADKFQSSNLYVKNLDPSISDEKLKEIFSPFGTVTSSKVMRDP-NGTSKGSGFV 363
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVV 87
FA P A + ++ L+G+ + + L V
Sbjct 364 AFATPEEATEAMSQLSGKMIESKPLYV 390
Score = 31.2 bits (69), Expect = 0.77, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 38/74 (51%), Gaps = 0/74 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T+LY+G++ + + + + G+ G V+ D ++ + +G+ +F P AA+ +
Sbjct 36 TSLYVGDLDFNVTDSQLFDAFGQMGTVVTVRVCRDLVTRRSLGYGYVNFTNPQDAARAIQ 95
Query 74 CLAGRKLHGRSLVV 87
L L+G+ + V
Sbjct 96 ELNYIPLYGKPIRV 109
Score = 31.2 bits (69), Expect = 0.87, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 35/75 (46%), Gaps = 1/75 (1%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
A ++I N+ + D+ + + FG ++ D SGQ +GF +A +A K +
Sbjct 123 AGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVAVDS-SGQSKGYGFVQYANEESAQKAI 181
Query 73 ACLAGRKLHGRSLVV 87
L G L+ + + V
Sbjct 182 EKLNGMLLNDKQVYV 196
> At4g13850
Length=158
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 42/76 (55%), Gaps = 0/76 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+T L+IG ++ D+ +R+ FG V+ D +G+ FGF +F + AA +
Sbjct 34 STKLFIGGLSWGTDDASLRDAFAHFGDVVDAKVIVDRETGRSRGFGFVNFNDEGAATAAI 93
Query 73 ACLAGRKLHGRSLVVN 88
+ + G++L+GR + VN
Sbjct 94 SEMDGKELNGRHIRVN 109
> Hs4557493
Length=577
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI A + ++++ E G V+ + D +G+ +GFC++ + A +
Sbjct 17 SVFVGNIPYEATEEQLKDIFSEVGPVVSFRLVYDRETGKPKGYGFCEYQDQETALSAMRN 76
Query 75 LAGRKLHGRSLVVN 88
L GR+ GR+L V+
Sbjct 77 LNGREFSGRALRVD 90
> At5g54580
Length=156
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 41/82 (50%), Gaps = 0/82 (0%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+ Q + Q + +T L++ ++K ++ +R +FG V +D +SG FGF
Sbjct 43 ESQTPARPQAEPSTNLFVSGLSKRTTSEGLRTAFAQFGEVADAKVVTDRVSGYSKGFGFV 102
Query 61 DFAEPLAAAKCVACLAGRKLHG 82
+A +AK +A + G+ L G
Sbjct 103 RYATLEDSAKGIAGMDGKFLDG 124
> Hs8922388
Length=759
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
L I N++ D ++ + +FG VL N P G++ FGF F L A K + +
Sbjct 116 LIIRNLSFKCSEDDLKTVFAQFGAVLEVNIPRKP-DGKMRGFGFVQFKNLLEAGKALKGM 174
Query 76 AGRKLHGRSLVVN 88
+++ GR++ V+
Sbjct 175 NMKEIKGRTVAVD 187
> At5g51300
Length=804
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T LYIG + ++D + NL FG ++ D ++G +GF +A+ A V
Sbjct 480 TNLYIGFLPPMLEDDGLINLFSSFGEIVMAKVIKDRVTGLSKGYGFVKYADVQMANTAVQ 539
Query 74 CLAGRKLHGRSLVV 87
+ G + GR+L V
Sbjct 540 AMNGYRFEGRTLAV 553
> CE16126
Length=336
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
++++GNI+ D +R++ + G VL D +G+ +GF +F + A +
Sbjct 19 SVFVGNISYDVSEDTIRSIFSKAGNVLSIKMVHDRETGKPKGYGFIEFPDIQTAEVAIRN 78
Query 75 LAGRKLHGRSLVVNC 89
L G +L GR L V+
Sbjct 79 LNGYELSGRILRVDS 93
> CE18908
Length=357
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 52/129 (40%), Gaps = 13/129 (10%)
Query 3 QQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDF 62
QQ +T LYI + + ++D +R + ++G + D + +GF DF
Sbjct 70 QQHHDSTPLSSTNLYIRGLMPNTNDDLLREMCSKYGNIASTKAIMDKATNNCKGYGFVDF 129
Query 63 AEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEK 122
P AAA V L E V+A++AK+ ++ T N + E+
Sbjct 130 ESPQAAAAAVDGL-------------NTEGVQAQMAKLQQQEQDPTNLYIANLPLDFTEQ 176
Query 123 EIEEEYKRL 131
+E E +
Sbjct 177 MLETELNKF 185
> 7293214
Length=416
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 39/72 (54%), Gaps = 0/72 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++IG + + ++D ++ LL FG++ +N D +G + FC++ + + +A L
Sbjct 209 IFIGGLPNYLNDDQVKELLLSFGKLRAFNLVKDAATGLSKGYAFCEYVDLSITDQSIAGL 268
Query 76 AGRKLHGRSLVV 87
G +L + L+V
Sbjct 269 NGMQLGDKKLIV 280
> Hs4757926
Length=524
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 0/73 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
LY+G++ + D +R + FGR+ D +G+ +GF F++ A K + L
Sbjct 252 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 311
Query 76 AGRKLHGRSLVVN 88
G +L GR + V
Sbjct 312 NGFELAGRPMKVG 324
> Hs4505575
Length=633
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 28/116 (24%), Positives = 54/116 (46%), Gaps = 4/116 (3%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q +Q + + + LY+ N+ D++ +R FG + + G+ FGF
Sbjct 278 EQMKQDRITRYQGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVMME--GGRSKGFGFV 335
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERI--LATMRAYKN 114
F+ P A K V + GR + + L V ++ + A +T + + +A++RA N
Sbjct 336 CFSSPEEATKAVTEMNGRIVATKPLYVALAQRKEERQAHLTNQYMQRMASVRAVPN 391
Score = 35.4 bits (80), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 4/79 (5%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +YI N + D++ +++L FG L +D SG+ FGF F A K V
Sbjct 191 TNVYIKNFGEDMDDERLKDL---FGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVD 246
Query 74 CLAGRKLHGRSLVVNCNEK 92
+ G++L+G+ + V +K
Sbjct 247 EMNGKELNGKQIYVGRAQK 265
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 29/118 (24%), Positives = 51/118 (43%), Gaps = 9/118 (7%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++I N+ K DN + + FG +L D + +GF F AA + + +
Sbjct 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSK--GYGFVHFETQEAAERAIEKM 158
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRLKD 133
G L+ R + V R K K E + A + + N + +++++E RLKD
Sbjct 159 NGMLLNDRKVFVG-----RFKSRKEREAELGARAKEFTNVYIKNFGEDMDDE--RLKD 209
> SPBC839.10
Length=695
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 29/53 (54%), Gaps = 1/53 (1%)
Query 10 QQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDF 62
QQ L+IGNI K D+ +M +L G++ W R +D + +FGF +F
Sbjct 135 QQMRRMLFIGNIPKELDDFWMDKILRLSGKLASWRRVAD-ADNSMTSFGFAEF 186
> At1g71800
Length=414
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 40/76 (52%), Gaps = 0/76 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
+++GNI A + +R + GE G V+ + +D +G+ +GFC++ + A L
Sbjct 11 VFVGNIPYDATEEQLREICGEVGPVVSFRLVTDRETGKPKGYGFCEYKDEETALSARRNL 70
Query 76 AGRKLHGRSLVVNCNE 91
+++GR L V+ E
Sbjct 71 QSYEINGRQLRVDFAE 86
> At3g08000
Length=143
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 1/81 (1%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
L+IG ++ D +++ FG V D SG+ FGF DFAE A +
Sbjct 43 LFIGGLSWSVDEQSLKDAFSSFGEVAEVRIAYDKGSGRSRGFGFVDFAEEGDALSAKDAM 102
Query 76 AGRKLHGRSLVVNCN-EKVRA 95
G+ L GR L ++ E+VR
Sbjct 103 DGKGLLGRPLRISFALERVRG 123
> At3g06970
Length=272
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 41/88 (46%), Gaps = 7/88 (7%)
Query 5 QQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAE 64
Q R T +++GN+T D +R +FG+V+ N S+ G+ +GF F +
Sbjct 3 QTNNHDTRVTKIFVGNLTWRTTADDLRRYFEQFGQVVDANVVSETYPGRSKGYGFITFRD 62
Query 65 PLAAAKCVACLAGRK--LHGRSLVVNCN 90
++ V L K + GR+ NCN
Sbjct 63 YVS---TVRALQNSKPIIDGRT--TNCN 85
> ECU08g0380
Length=214
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 39/82 (47%), Gaps = 1/82 (1%)
Query 10 QQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAA 69
+ ++++GNI + + LG G+V+ + D +G+ FGFC++ PL
Sbjct 1 MKSGCSIFVGNIDFEVPEERIIEELGAVGKVVSFRMVYDKTTGKSRGFGFCEYESPLIVE 60
Query 70 KCVACLAGRKLHGRSLVVNCNE 91
K + L +GR + +N E
Sbjct 61 KALQNLK-ISFNGRPVKINYAE 81
> 7299786
Length=690
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 6/64 (9%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVL------RWNRQSDPISGQLAAFGFCDFAEPLAAA 69
L++GNI HA D +R + FG VL + + P +GF + +P A
Sbjct 495 LFLGNIPHHASEDDLREIFSRFGNVLELRILSKAGNKVPPGMRSPLNYGFITYDDPEAVQ 554
Query 70 KCVA 73
KC+A
Sbjct 555 KCLA 558
> At2g23350
Length=662
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 26/118 (22%), Positives = 53/118 (44%), Gaps = 9/118 (7%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+++++ + + T +Y+ N+++ +D ++ G++G + D G+ FGF
Sbjct 212 KEERESAADKMKFTNVYVKNLSEATTDDELKTTFGQYGSISSAVVMRDG-DGKSRCFGFV 270
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTRE 118
+F P AA+ V L G+K + V +K ER L R Y+ + +
Sbjct 271 NFENPEDAARAVEALNGKKFDDKEWYVGKAQK--------KSERELELSRRYEQGSSD 320
Score = 35.8 bits (81), Expect = 0.036, Method: Composition-based stats.
Identities = 22/88 (25%), Positives = 41/88 (46%), Gaps = 1/88 (1%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
LY+ N+ ++ +R L EFG + DP SG GF F+ A++ + +
Sbjct 330 LYVKNLDDTVTDEKLRELFAEFGTITSCKVMRDP-SGTSKGSGFVAFSAASEASRVLNEM 388
Query 76 AGRKLHGRSLVVNCNEKVRAKIAKVTEE 103
G+ + G+ L V ++ + AK+ +
Sbjct 389 NGKMVGGKPLYVALAQRKEERRAKLQAQ 416
> At5g47320
Length=244
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/77 (23%), Positives = 40/77 (51%), Gaps = 0/77 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+T LYIG ++ D +++ F V ++ ++G+ +GF +F +A +
Sbjct 62 STKLYIGGLSPGTDEHSLKDAFSSFNGVTEARVMTNKVTGRSRGYGFVNFISEDSANSAI 121
Query 73 ACLAGRKLHGRSLVVNC 89
+ + G++L+G ++ VN
Sbjct 122 SAMNGQELNGFNISVNV 138
> SPAC140.02
Length=500
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 1/76 (1%)
Query 8 QQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLA 67
Q + + T+++GN++ +A D + G G + +DP SG+L FG+ F++ +
Sbjct 360 QLSEPSDTVFVGNLSFNATEDDLSTAFGGCGDIQSIRLPTDPQSGRLKGFGYVTFSDIDS 419
Query 68 AAKCVACLAGRKLHGR 83
A KCV + G + GR
Sbjct 420 AKKCVE-MNGHFIAGR 434
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 13/58 (22%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 8 QQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEP 65
Q T+++G ++ + D+ ++ E+G ++ D SG+ +G+ DF P
Sbjct 257 QDSNETCTVFVGRLSWNVDDQWLGQEFEEYGTIVGARVIMDGQSGRSKGYGYVDFETP 314
> CE18909
Length=409
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 50/119 (42%), Gaps = 13/119 (10%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+T LYI + + ++D +R + ++G + D + +GF DF P AAA V
Sbjct 106 STNLYIRGLMPNTNDDLLREMCSKYGNIASTKAIMDKATNNCKGYGFVDFESPQAAAAAV 165
Query 73 ACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEKEIEEEYKRL 131
L E V+A++AK+ ++ T N + E+ +E E +
Sbjct 166 DGL-------------NTEGVQAQMAKLQQQEQDPTNLYIANLPLDFTEQMLETELNKF 211
> Hs20559158
Length=330
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +Y+ N+ D +++L +FG++L D SG FGF +F + A K V
Sbjct 191 TNIYVKNLPVDVDEQGLQDLFSQFGKMLSVKVMRDN-SGHSRCFGFVNFEKHEEAQKAVV 249
Query 74 CLAGRKLHGRSL 85
+ G+++ GR L
Sbjct 250 HMNGKEVSGRLL 261
> CE20412
Length=646
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 56/117 (47%), Gaps = 5/117 (4%)
Query 1 QQQQQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFC 60
+Q + ++ Q+ + LY+ N+ + D+D ++ +G + +D +G+ FGF
Sbjct 304 EQHKAERMQKYQGVNLYVKNLDETVDDDGLKKQFESYGNITSAKVMTDE-NGRSKGFGFV 362
Query 61 DFAEPLAAAKCVACLAGRKLHGRSLVVNC---NEKVRAKIAKVTEERILATMRAYKN 114
F +P A V + + + + L V E RA++A +R LA+MR + N
Sbjct 363 CFEKPEEATSAVTEMNSKMVCSKPLYVAIAQRKEDRRAQLASQYMQR-LASMRMHGN 418
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 30/72 (41%), Gaps = 2/72 (2%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T +Y+ N H + + + L +FG + + + G+ FGF FA P A V
Sbjct 213 TNVYVKNFGDHYNKETLEKLFAKFGNITSCEVMT--VEGKSKGFGFVAFANPEEAETAVQ 270
Query 74 CLAGRKLHGRSL 85
L + G L
Sbjct 271 ALHDSTIEGTDL 282
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 31/75 (41%), Gaps = 1/75 (1%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
A ++I N+ K DN + + FG +L D G +GF F AA +
Sbjct 119 AGNIFIKNLDKVIDNKSIYDTFSLFGNILSCKVAIDE-DGFSKGYGFVHFETEEAAQNAI 177
Query 73 ACLAGRKLHGRSLVV 87
+ G L G+ + V
Sbjct 178 QKVNGMLLAGKKVFV 192
> CE02193
Length=692
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 53/110 (48%), Gaps = 5/110 (4%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
+Q++ Q+ + LY+ NI + ++D +R+ FG + D +G+ FGF F
Sbjct 333 KQERVQRYQGVNLYVKNIEEDLNDDGLRDHFSSFGTITSAKVMVDE-NGRSKGFGFVCFE 391
Query 64 EPLAAAKCVACLAGRKLHGRSLVVNC---NEKVRAKIAKVTEERILATMR 110
+P A V + + + + L V E RA++A +R LAT+R
Sbjct 392 KPEEATAAVTDMNSKMIGAKPLYVALAQRKEDRRAQLASQYMQR-LATLR 440
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 33/71 (46%), Gaps = 1/71 (1%)
Query 12 RATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKC 71
+ T +++ N +H D + + + +FG + +D G+ FGF FA+ AA +
Sbjct 236 KYTNVFVKNFGEHLDQEKLSAMFSKFGEITSAVVMTD-AQGKPKGFGFVAFADQDAAGQA 294
Query 72 VACLAGRKLHG 82
V L L G
Sbjct 295 VEKLNDSILEG 305
> SPAC644.16
Length=422
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+Y+GNI + + ++ + G V + DP SGQ +GFC++ +P AA V
Sbjct 7 VYVGNIPYEMAEEQVIDIFKQSGPVKSFQLVIDPESGQPKGYGFCEYHDPATAASAV 63
> Hs6005926
Length=475
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 39/75 (52%), Gaps = 0/75 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
A L+IG + + ++D ++ LL FG + +N D +G + FC++ + + +
Sbjct 258 AHKLFIGGLPNYLNDDQVKELLTSFGPLKAFNLVKDSATGLSKGYAFCEYVDINVTDQAI 317
Query 73 ACLAGRKLHGRSLVV 87
A L G +L + L+V
Sbjct 318 AGLNGMQLGDKKLLV 332
> CE03230
Length=566
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 53/110 (48%), Gaps = 5/110 (4%)
Query 4 QQQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFA 63
+Q++ Q+ + LY+ NI + ++D +R+ FG + D +G+ FGF F
Sbjct 333 KQERVQRYQGVNLYVKNIEEDLNDDGLRDHFSSFGTITSAKVMVDE-NGRSKGFGFVCFE 391
Query 64 EPLAAAKCVACLAGRKLHGRSLVVNC---NEKVRAKIAKVTEERILATMR 110
+P A V + + + + L V E RA++A +R LAT+R
Sbjct 392 KPEEATAAVTDMNSKMIGAKPLYVALAQRKEDRRAQLASQYMQR-LATLR 440
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 50/111 (45%), Gaps = 7/111 (6%)
Query 12 RATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKC 71
+ T +++ N +H D + + + +FG + +D G+ FGF FA+ AA +
Sbjct 236 KYTNVFVKNFGEHLDQEKLSAMFSKFGEITSAVVMTD-AQGKPKGFGFVAFADQDAAGQA 294
Query 72 VACLAGRKLHGRSLVVNCNEKVRAKIAKVTEERILATMRAYKNKTREQVEK 122
V L L G +C K+ A+ ER R Y+ +E+V++
Sbjct 295 VEKLNDSILEG----TDC--KLSVCRAQKKSERSAELKRKYEALKQERVQR 339
> YGR159c
Length=414
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 36/73 (49%), Gaps = 0/73 (0%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCV 72
+ TL++GN++ +AD D + L + G V+ + P + Q FG+ F+ A K +
Sbjct 266 SDTLFLGNLSFNADRDAIFELFAKHGEVVSVRIPTHPETEQPKGFGYVQFSNMEDAKKAL 325
Query 73 ACLAGRKLHGRSL 85
L G + R +
Sbjct 326 DALQGEYIDNRPV 338
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 19/92 (20%), Positives = 49/92 (53%), Gaps = 10/92 (10%)
Query 3 QQQQQQQQQRATTLYIGNITKHADNDFMR----NLLGEFGRVLRWNRQSDPISGQLAAFG 58
++Q+ ++ + T+++G ++ D+++++ ++ G G + + R +D G +G
Sbjct 157 KKQKNEETEEPATIFVGRLSWSIDDEWLKKEFEHIGGVIGARVIYERGTDRSRG----YG 212
Query 59 FCDFAEPLAAAKCVACLAGRKLHGRSLVVNCN 90
+ DF A K + + G+++ GR +NC+
Sbjct 213 YVDFENKSYAEKAIQEMQGKEIDGRP--INCD 242
> Hs5729884
Length=556
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 49/87 (56%), Gaps = 15/87 (17%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQL---AAFGFCDFAEPLAAAKCV 72
LYIGN++ D +R L G+ R L P++GQ+ + + F D+ + A + +
Sbjct 5 LYIGNLSPAVTADDLRQLFGD--RKL-------PLAGQVLLKSGYAFVDYPDQNWAIRAI 55
Query 73 ACLAGR-KLHGRSLVVN--CNEKVRAK 96
L+G+ +LHG+ + V+ ++K+R++
Sbjct 56 ETLSGKVELHGKIMEVDYSVSKKLRSR 82
> At3g53460
Length=342
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 0/73 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
LY+GN++ D+ + NL E G+V+ D SG+ FGF + K + L
Sbjct 259 LYVGNLSWGVDDMALENLFNEQGKVVEARVIYDRDSGRSKGFGFVTLSSSQEVQKAINSL 318
Query 76 AGRKLHGRSLVVN 88
G L GR + V+
Sbjct 319 NGADLDGRQIRVS 331
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 18/74 (24%), Positives = 30/74 (40%), Gaps = 0/74 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
L++GN++ + D+ + L G V D ++G+ FGF +
Sbjct 101 LFVGNLSFNVDSAQLAQLFESAGNVEMVEVIYDKVTGRSRGFGFVTMSTAAEVEAAAQQF 160
Query 76 AGRKLHGRSLVVNC 89
G + GR L VN
Sbjct 161 NGYEFEGRPLRVNA 174
> At2g18510
Length=363
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 26/116 (22%), Positives = 45/116 (38%), Gaps = 13/116 (11%)
Query 13 ATTLYIGNITKHADNDFMRNLLGEFG------RVLRWNRQSDPISGQLAAFGFCDFAEPL 66
L+IGN+ D + + FG +++R DP +G FGF +
Sbjct 111 GANLFIGNLDPDVDEKLLYDTFSAFGVIASNPKIMR-----DPDTGNSRGFGFISYDSFE 165
Query 67 AAAKCVACLAGRKLHGRSLVVNCNEK--VRAKIAKVTEERILATMRAYKNKTREQV 120
A+ + + G+ L R + V+ K + + ER+LA K+R
Sbjct 166 ASDAAIESMTGQYLSNRQITVSYAYKKDTKGERHGTPAERLLAATNPTAQKSRPHT 221
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/84 (21%), Positives = 36/84 (42%), Gaps = 0/84 (0%)
Query 5 QQQQQQQRATTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAE 64
Q ++ + T+Y+G + + + L + G V+ D ++ +GF ++
Sbjct 16 QHSAERNQDATVYVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRS 75
Query 65 PLAAAKCVACLAGRKLHGRSLVVN 88
A + L KLHG+ + VN
Sbjct 76 EEDADYAIKVLNMIKLHGKPIRVN 99
> SPBC4F6.14
Length=674
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 34/72 (47%), Gaps = 0/72 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
+TL++ N+ D + N + G + ++P +G+ +GF F+ A +
Sbjct 5 STLFVRNLAFQTKQDDLTNFFSDVGPIKHAVVVTNPETGENRGYGFVTFSMLEDAQRAAK 64
Query 74 CLAGRKLHGRSL 85
L +KLHGR L
Sbjct 65 ELKNKKLHGRIL 76
> CE02197
Length=256
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 0/73 (0%)
Query 15 TLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVAC 74
T + N+ + + D +R+L G+ GRV+R D ++G F F F AA+ +A
Sbjct 177 TCRVTNLPQEMNEDELRDLFGKIGRVIRIFIARDKVTGLPKGFAFVTFESRDDAARAIAE 236
Query 75 LAGRKLHGRSLVV 87
L +++ L V
Sbjct 237 LNDIRMYHMVLKV 249
> 7294907
Length=918
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 31/70 (44%), Gaps = 0/70 (0%)
Query 16 LYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVACL 75
++ N+ + +R L +FG V+ N D ++ ++ FG + P A K L
Sbjct 366 IFFRNLAYTTTEEDLRKLFEQFGPVVEVNLPLDKLTRKIKGFGTVTYMMPEHALKAFNTL 425
Query 76 AGRKLHGRSL 85
G HGR L
Sbjct 426 DGTDFHGRLL 435
> Hs11386163
Length=380
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 0/84 (0%)
Query 14 TTLYIGNITKHADNDFMRNLLGEFGRVLRWNRQSDPISGQLAAFGFCDFAEPLAAAKCVA 73
T L + + ++ + R+L G G + D I+GQ +GF ++ +P A K +
Sbjct 46 TNLIVNYLPQNMTQEEFRSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAIN 105
Query 74 CLAGRKLHGRSLVVNCNEKVRAKI 97
L G +L +++ V+ A I
Sbjct 106 TLNGLRLQTKTIKVSYARPSSASI 129
Lambda K H
0.316 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40