bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2980_orf2
Length=104
Score E
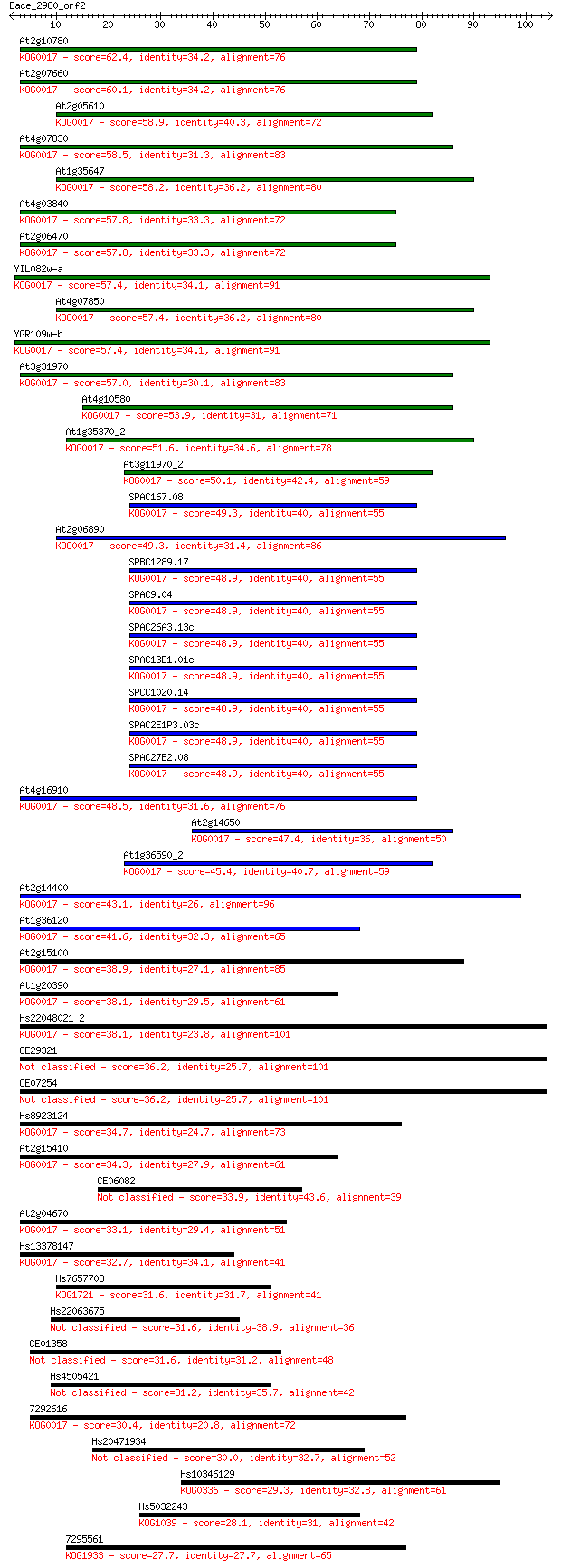
Sequences producing significant alignments: (Bits) Value
At2g10780 62.4 2e-10
At2g07660 60.1 9e-10
At2g05610 58.9 2e-09
At4g07830 58.5 3e-09
At1g35647 58.2 4e-09
At4g03840 57.8 5e-09
At2g06470 57.8 5e-09
YIL082w-a 57.4 6e-09
At4g07850 57.4 6e-09
YGR109w-b 57.4 6e-09
At3g31970 57.0 9e-09
At4g10580 53.9 8e-08
At1g35370_2 51.6 3e-07
At3g11970_2 50.1 1e-06
SPAC167.08 49.3 2e-06
At2g06890 49.3 2e-06
SPBC1289.17 48.9 2e-06
SPAC9.04 48.9 2e-06
SPAC26A3.13c 48.9 2e-06
SPAC13D1.01c 48.9 2e-06
SPCC1020.14 48.9 2e-06
SPAC2E1P3.03c 48.9 2e-06
SPAC27E2.08 48.9 2e-06
At4g16910 48.5 3e-06
At2g14650 47.4 7e-06
At1g36590_2 45.4 2e-05
At2g14400 43.1 1e-04
At1g36120 41.6 4e-04
At2g15100 38.9 0.002
At1g20390 38.1 0.004
Hs22048021_2 38.1 0.004
CE29321 36.2 0.014
CE07254 36.2 0.014
Hs8923124 34.7 0.041
At2g15410 34.3 0.059
CE06082 33.9 0.081
At2g04670 33.1 0.14
Hs13378147 32.7 0.18
Hs7657703 31.6 0.35
Hs22063675 31.6 0.38
CE01358 31.6 0.41
Hs4505421 31.2 0.51
7292616 30.4 0.93
Hs20471934 30.0 1.0
Hs10346129 29.3 1.9
Hs5032243 28.1 4.4
7295561 27.7 5.4
> At2g10780
Length=1611
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 0/76 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + H G K + L + Y+W GM+ +V C C+ K +Q P+G LQ L I
Sbjct 1168 HQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPI 1227
Query 63 PSRRWSYVSLDFITDL 78
P +W ++++DF+T L
Sbjct 1228 PEWKWDHITMDFVTGL 1243
> At2g07660
Length=949
Score = 60.1 bits (144), Expect = 9e-10, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 40/76 (52%), Gaps = 0/76 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + H G K + L + Y+W GMR +V C C+ K +Q P+G LQ L I
Sbjct 560 HQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPI 619
Query 63 PSRRWSYVSLDFITDL 78
+W ++++DF+T L
Sbjct 620 SEWKWDHITMDFVTRL 635
> At2g05610
Length=780
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 10 HRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSY 69
H G++ T + L+YW M A++ SC C+ K N G LQ L IP R WS
Sbjct 487 HSGKEVTHQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWSD 546
Query 70 VSLDFITDLQLT 81
VS+DFI L L+
Sbjct 547 VSMDFIDGLPLS 558
> At4g07830
Length=611
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 26/83 (31%), Positives = 43/83 (51%), Gaps = 0/83 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + + H G K + L + Y W GM+ +VE C C+ K +Q LQ L I
Sbjct 178 HASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEECDVCQLVKIEHQVSGSLLQSLPI 237
Query 63 PSRRWSYVSLDFITDLQLTKTGD 85
P +W ++++DF+ L +++T D
Sbjct 238 PEWKWDFITMDFVVGLPVSRTKD 260
> At1g35647
Length=1495
Score = 58.2 bits (139), Expect = 4e-09, Method: Composition-based stats.
Identities = 29/80 (36%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Query 10 HRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSY 69
H G KT + ++WP M+ E C C+ +K +Q P G L IPS W+
Sbjct 1007 HFGVSKTIKVMQDHFHWPHMKRDVERICERCPTCKQAKAKSQ-PHGLYTPLPIPSHPWND 1065
Query 70 VSLDFITDLQLTKTGDDSIL 89
+S+DF+ L T+TG DSI
Sbjct 1066 ISMDFVVGLPRTRTGKDSIF 1085
> At4g03840
Length=973
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + H G K + L + Y+W GM+ +V C C+ K +Q P+G LQ L I
Sbjct 618 HQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPI 677
Query 63 PSRRWSYVSLDF 74
P +W ++++DF
Sbjct 678 PEWKWDHITMDF 689
> At2g06470
Length=899
Score = 57.8 bits (138), Expect = 5e-09, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + H G K + L + Y+W GM+ +V C C+ K +Q P+G LQ L I
Sbjct 618 HQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPI 677
Query 63 PSRRWSYVSLDF 74
P +W ++++DF
Sbjct 678 PEWKWDHITMDF 689
> YIL082w-a
Length=1498
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 46/92 (50%), Gaps = 1/92 (1%)
Query 2 HHDH-VTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQL 60
+HDH + H G T +S +YYWP ++ Y+ +C QC+ K + G LQ L
Sbjct 1129 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1188
Query 61 IIPSRRWSYVSLDFITDLQLTKTGDDSILCWL 92
I RW +S+DF+T L T + IL +
Sbjct 1189 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVV 1220
> At4g07850
Length=1138
Score = 57.4 bits (137), Expect = 6e-09, Method: Composition-based stats.
Identities = 29/80 (36%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Query 10 HRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSY 69
H G KT + ++WP M+ E CT C+ +K +Q P G L IP W+
Sbjct 774 HFGVSKTLKVMQDHFHWPHMKRDVERMCERCTTCKQAKAKSQ-PHGLCTPLPIPLHPWND 832
Query 70 VSLDFITDLQLTKTGDDSIL 89
+S+DF+ L T+TG DSI
Sbjct 833 ISMDFVVGLPRTRTGKDSIF 852
> YGR109w-b
Length=1547
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 46/92 (50%), Gaps = 1/92 (1%)
Query 2 HHDH-VTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQL 60
+HDH + H G T +S +YYWP ++ Y+ +C QC+ K + G LQ L
Sbjct 1103 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1162
Query 61 IIPSRRWSYVSLDFITDLQLTKTGDDSILCWL 92
I RW +S+DF+T L T + IL +
Sbjct 1163 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVV 1194
> At3g31970
Length=1329
Score = 57.0 bits (136), Expect = 9e-09, Method: Composition-based stats.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + + H K + L + Y W GM+ +V C C+ K +Q P G LQ L I
Sbjct 911 HASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPI 970
Query 63 PSRRWSYVSLDFITDLQLTKTGD 85
+W ++++DF+ L +++T D
Sbjct 971 LEWKWDFITMDFVVGLPVSRTKD 993
> At4g10580
Length=1240
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 38/71 (53%), Gaps = 0/71 (0%)
Query 15 KTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDF 74
K + L + Y W GM+ +V C C+ K +Q G LQ L IP +W ++++D
Sbjct 960 KMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDL 1019
Query 75 ITDLQLTKTGD 85
+ L++++T D
Sbjct 1020 VVGLRVSRTKD 1030
> At1g35370_2
Length=923
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 39/78 (50%), Gaps = 2/78 (2%)
Query 12 GQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVS 71
G+ + + L+YW GM A++ SC C+ K N G LQ L IP + W VS
Sbjct 545 GRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIWCDVS 604
Query 72 LDFITDLQLTKTGDDSIL 89
+DFI L +G S++
Sbjct 605 MDFIEG--LPNSGGKSVI 620
> At3g11970_2
Length=958
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 23 LYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDLQLT 81
L+YW GM AY+ SC C+ K G LQ L IP WS VS+DFI L ++
Sbjct 568 LFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVS 626
> SPAC167.08
Length=1214
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 822 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 876
> At2g06890
Length=1215
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/86 (31%), Positives = 41/86 (47%), Gaps = 3/86 (3%)
Query 10 HRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSY 69
H G KT +++ + WP M+ C C+ +K Q P G L IP W+
Sbjct 908 HFGIAKTLEVMTEHFRWPHMKCDVKRICGRCNTCKQAKSKIQ-PNGLYTPLPIPKHPWND 966
Query 70 VSLDFITDLQLTKTGDDSILCWLTPL 95
+S+DF+ + L +TG DSI +P
Sbjct 967 ISMDFV--MGLPRTGKDSIFVVYSPF 990
> SPBC1289.17
Length=1333
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 941 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 995
> SPAC9.04
Length=1333
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 941 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 995
> SPAC26A3.13c
Length=1333
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 941 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 995
> SPAC13D1.01c
Length=1333
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 941 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 995
> SPCC1020.14
Length=1333
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 941 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 995
> SPAC2E1P3.03c
Length=1333
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 941 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 995
> SPAC27E2.08
Length=1333
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 24 YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDL 78
+ W G+R YV++C C+ +K N KP G LQ + R W +S+DFIT L
Sbjct 941 FTWKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL 995
> At4g16910
Length=687
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 38/76 (50%), Gaps = 8/76 (10%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + H G K + L + Y+W GM+ +V +K +Q P+G LQ L I
Sbjct 260 HQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV--------AKEEHQVPSGMLQNLPI 311
Query 63 PSRRWSYVSLDFITDL 78
P +W ++ +DF+T L
Sbjct 312 PEWKWDHIMMDFVTGL 327
> At2g14650
Length=1328
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 36 YVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDLQLTKTGD 85
+V C C+ K +Q P G LQ L IP +W ++++DF+ L +++T D
Sbjct 946 WVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTKD 995
> At1g36590_2
Length=958
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 30/59 (50%), Gaps = 0/59 (0%)
Query 23 LYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDLQLT 81
L+Y GM AY+ SC C+ K G LQ L IP WS VS+DFI L ++
Sbjct 568 LFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVS 626
> At2g14400
Length=1466
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/99 (25%), Positives = 40/99 (40%), Gaps = 6/99 (6%)
Query 3 HDHVTARHRGQKKTFLSLSKL-YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLI 61
H+ + H + + K+ YYWP M T Y + C +C+ PL +P+ +
Sbjct 979 HEGLCGSHSSGRAMAFKIKKMCYYWPTMITDCVKYAQRCKRCQLHAPLIHQPSELFSSIS 1038
Query 62 IPS--RRWSYVSLDFITDLQLTKTGDDSILCWLTPLVRW 98
P RW S+D I L + G +L +W
Sbjct 1039 APYPFMRW---SMDIIGPLHRSTRGVQYLLVLTDYFSKW 1074
> At1g36120
Length=1235
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 29/65 (44%), Gaps = 2/65 (3%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H + + H G K + L + Y W GM+ +V C C+ K +Q P G LQ L
Sbjct 884 HASMFSIHPGATKMYRDLKRHYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSL-- 941
Query 63 PSRRW 67
P W
Sbjct 942 PISEW 946
> At2g15100
Length=1329
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 37/86 (43%), Gaps = 2/86 (2%)
Query 3 HDHVTARHRGQKKTFLSLSKL-YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLI 61
HD H G + + K YYWP + AY C QC+ PL +PA L +
Sbjct 994 HDGTCGNHTGGRSLAFKVRKYGYYWPTLVADCEAYARKCEQCQKHAPLILQPAELLTTVS 1053
Query 62 IPSRRWSYVSLDFITDLQLTKTGDDS 87
P ++ +D + L ++ G ++
Sbjct 1054 APYPFMKWL-MDIVGPLHVSTRGVEA 1078
> At1g20390
Length=1791
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 27/62 (43%), Gaps = 1/62 (1%)
Query 3 HDHVTARHRGQKKTFLSLSKL-YYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLI 61
H+ H G + L L KL +YWP M + + C QC+ P +P L+ +
Sbjct 1443 HEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTELLRAGV 1502
Query 62 IP 63
P
Sbjct 1503 AP 1504
> Hs22048021_2
Length=835
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/106 (22%), Positives = 46/106 (43%), Gaps = 9/106 (8%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
HD H+ ++T+ L L +WPGM+ + Y SC C + + L+ +
Sbjct 486 HDIPLGAHQRPEETYKKLRLLGWWPGMQEHVKDYCRSCLFCIPRNLIGSE----LKVIES 541
Query 63 P-----SRRWSYVSLDFITDLQLTKTGDDSILCWLTPLVRWHTSSP 103
P + WS + ++ + + +++ G +L P RW + P
Sbjct 542 PWPLRSTAPWSNLQIEVVGPVTISEEGHKHVLIVADPNTRWVEAFP 587
> CE29321
Length=2186
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 9/104 (8%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H+ + A H G KK + + + +YWP MR V +C +C + ++ L +
Sbjct 1473 HEGMLAGHFGIKKMWRMVHRKFYWPQMRVCVENCVRTCAKCLCANDHSK-----LTSSLT 1527
Query 63 PSRR---WSYVSLDFITDLQLTKTGDDSILCWLTPLVRWHTSSP 103
P R V+ D + D+ L+ G+ IL + ++ T+ P
Sbjct 1528 PYRMTFPLEIVACDLM-DVGLSVQGNRYILTIIDLFTKYGTAVP 1570
> CE07254
Length=2175
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 9/104 (8%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H+ + A H G KK + + + +YWP MR V +C +C + ++ L +
Sbjct 1462 HEGMLAGHFGIKKMWRMVHRKFYWPQMRVCVENCVRTCAKCLCANDHSK-----LTSSLT 1516
Query 63 PSRR---WSYVSLDFITDLQLTKTGDDSILCWLTPLVRWHTSSP 103
P R V+ D + D+ L+ G+ IL + ++ T+ P
Sbjct 1517 PYRMTFPLEIVACDLM-DVGLSVQGNRYILTIIDLFTKYGTAVP 1559
> Hs8923124
Length=392
Score = 34.7 bits (78), Expect = 0.041, Method: Composition-based stats.
Identities = 18/73 (24%), Positives = 32/73 (43%), Gaps = 2/73 (2%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLII 62
H++ + H G +T + YYW + +V +C C+ +K N Q L+
Sbjct 78 HENDSGAHHGISRTLTLVESNYYWTSVTNDAKQWVYACQHCQVAK--NTVIVAPKQHLLK 135
Query 63 PSRRWSYVSLDFI 75
WS V++D +
Sbjct 136 VENPWSLVTVDLM 148
> At2g15410
Length=1787
Score = 34.3 bits (77), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 25/62 (40%), Gaps = 1/62 (1%)
Query 3 HDHVTARHRGQKKTFLSLSK-LYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLI 61
H+ H G + L + K YWP M +V C +C+ P P+ LQ +
Sbjct 1439 HEGAGGNHSGGRALALKIRKHGQYWPTMNADCEKFVARCEKCQGHAPFIHIPSEVLQTAM 1498
Query 62 IP 63
P
Sbjct 1499 PP 1500
> CE06082
Length=172
Score = 33.9 bits (76), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Query 18 LSLSKLYYWPGMRTYTTAYVESCTQ--CRASKPLNQKPAGF 56
L +S Y P +RT T Y T RA P+NQ+P GF
Sbjct 82 LEVSSREYDPPIRTTTPEYFPPVTATIVRAEGPINQRPIGF 122
> At2g04670
Length=1411
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKP 53
H + + H G K + L + Y W GM+ +V C C+ K +Q P
Sbjct 1024 HASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWVAECDVCQLVKAEHQVP 1074
> Hs13378147
Length=150
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 3 HDHVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQC 43
HD H+ ++T+ L L +WPGM+ + Y SC C
Sbjct 78 HDIPLGAHQRPEETYKKLRLLGWWPGMQEHVKDYCRSCLFC 118
> Hs7657703
Length=1053
Score = 31.6 bits (70), Expect = 0.35, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 26/45 (57%), Gaps = 4/45 (8%)
Query 10 HRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRA----SKPLN 50
H + +T+ LSK Y+W G+ Y++ C++C+ S+P++
Sbjct 86 HHTRHQTWHYLSKTYWWRGILKQVKDYIKQCSKCQEKLDRSRPIS 130
> Hs22063675
Length=163
Score = 31.6 bits (70), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 25/37 (67%), Gaps = 3/37 (8%)
Query 9 RHRGQKKTFLSLSKLY-YWPGMRTYTTAYVESCTQCR 44
+H GQKKT+ ++++ Y + P R T ++ SCT+C+
Sbjct 55 KHAGQKKTYRAIAETYAFLP--REAVTRFLMSCTECQ 89
> CE01358
Length=558
Score = 31.6 bits (70), Expect = 0.41, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 5 HVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQK 52
H H GQK+T + + + YW +R ++ SC C A K +K
Sbjct 122 HELIGHLGQKRTQMVVLRKLYWRSVRQDVKTFIASCDFCTAKKIQGRK 169
> Hs4505421
Length=524
Score = 31.2 bits (69), Expect = 0.51, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 27/43 (62%), Gaps = 3/43 (6%)
Query 9 RHRGQKKTFLSLSKLY-YWPGMRTYTTAYVESCTQCRASKPLN 50
+H GQK+T+ ++S+ Y + P R T ++ SC++C+ LN
Sbjct 14 KHAGQKRTYKAISESYAFLP--REAVTRFLMSCSECQKRMHLN 54
> 7292616
Length=1062
Score = 30.4 bits (67), Expect = 0.93, Method: Composition-based stats.
Identities = 15/72 (20%), Positives = 29/72 (40%), Gaps = 0/72 (0%)
Query 5 HVTARHRGQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPS 64
H+ HRG +T + + Y+P ++ + + C C+ K Q Q P+
Sbjct 738 HIKNNHRGIDETVSHIKRQIYFPCLKERVSQLINKCDICQTLKYDRQPQKPIFQLTETPN 797
Query 65 RRWSYVSLDFIT 76
+ V +D +
Sbjct 798 KPLDIVHIDLYS 809
> Hs20471934
Length=1028
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 8/52 (15%)
Query 17 FLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWS 68
F++L ++ M Y+T ++ S PLN+K A F ++L P RRW+
Sbjct 880 FMALDCVFQALTMEEYSTNWL--------SDPLNEKMAFFHEKLSAPERRWN 923
> Hs10346129
Length=648
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 27/61 (44%), Gaps = 1/61 (1%)
Query 34 TAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVSLDFITDLQLTKTGDDSILCWLT 93
T + QC N K AGF + I S+ W V L I + + +TG LC+L
Sbjct 240 TCTFDDAFQCYPEVMENIKKAGFQKPTPIQSQAWPIV-LQGIDLIGVAQTGTGKTLCYLM 298
Query 94 P 94
P
Sbjct 299 P 299
> Hs5032243
Length=507
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 18/42 (42%), Gaps = 11/42 (26%)
Query 26 WPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRW 67
W R + V+SC QCR + +L+IPS W
Sbjct 347 WRSARQFENRIVKSCPQCRVT-----------SELVIPSEFW 377
> 7295561
Length=1223
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 24/65 (36%), Gaps = 10/65 (15%)
Query 12 GQKKTFLSLSKLYYWPGMRTYTTAYVESCTQCRASKPLNQKPAGFLQQLIIPSRRWSYVS 71
G + +L LS L +Y +Y +C C S PL P G P W
Sbjct 187 GSNEIYLDLSPL---KCGESYEDSYACACIDCEESCPLTDAPTG-------PDELWKIAG 236
Query 72 LDFIT 76
L +T
Sbjct 237 LYGVT 241
Lambda K H
0.322 0.133 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40