bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_2991_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
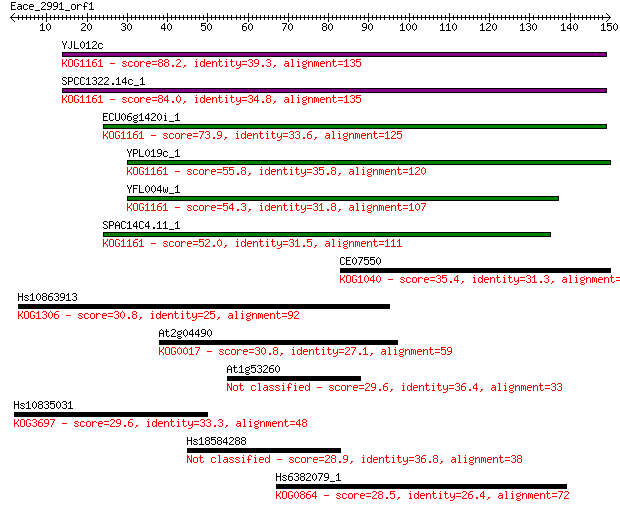
YJL012c 88.2 5e-18
SPCC1322.14c_1 84.0 1e-16
ECU06g1420i_1 73.9 1e-13
YPL019c_1 55.8 3e-08
YFL004w_1 54.3 7e-08
SPAC14C4.11_1 52.0 4e-07
CE07550 35.4 0.040
Hs10863913 30.8 1.0
At2g04490 30.8 1.0
At1g53260 29.6 2.1
Hs10835031 29.6 2.4
Hs18584288 28.9 4.1
Hs6382079_1 28.5 5.4
> YJL012c
Length=648
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 53/135 (39%), Positives = 74/135 (54%), Gaps = 24/135 (17%)
Query 14 ESEAWKPPDSFVRVTTKYWVRPENIVRAQCLIVRHLPFLVFGTSDKDLEASLLGPEATAN 73
+S A +FVR TTKYWV P+NI + +I++HLP LVF T +K+ E E +A
Sbjct 184 DSSAGGKQQNFVRQTTKYWVHPDNITELKLIILKHLPVLVFNT-NKEFER-----EDSA- 236
Query 74 GHNFRGPNLAPTQMVASVYFDSSSAYSYERRIRRFEGAQLLRFRWYGTNNDGPDEDIFIE 133
+ S+YFD+ + Y R+R+ EGA+ R RWYG + + IF+E
Sbjct 237 --------------ITSIYFDNENLDLYYGRLRKDEGAEAHRLRWYGGMS---TDTIFVE 279
Query 134 RKTHHESWSGMSSTK 148
RKTH E W+G S K
Sbjct 280 RKTHREDWTGEKSVK 294
> SPCC1322.14c_1
Length=608
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 47/135 (34%), Positives = 68/135 (50%), Gaps = 24/135 (17%)
Query 14 ESEAWKPPDSFVRVTTKYWVRPENIVRAQCLIVRHLPFLVFGTSDKDLEASLLGPEATAN 73
+S A +FVR TTKYWV P N+ + I++HLP LVF + + E A
Sbjct 182 DSAAGGTQQNFVRQTTKYWVHPNNVTELKIYILKHLPVLVFNPNKE------FAREDAA- 234
Query 74 GHNFRGPNLAPTQMVASVYFDSSSAYSYERRIRRFEGAQLLRFRWYGTNNDGPDEDIFIE 133
+ S+Y+D+ Y R+ + EGA+ +R RWYG ++ +IF+E
Sbjct 235 --------------ITSIYYDNDDLDFYLGRLEKREGAEAIRLRWYGNMDNN---NIFVE 277
Query 134 RKTHHESWSGMSSTK 148
RKTH E W+G S K
Sbjct 278 RKTHREDWTGEKSVK 292
> ECU06g1420i_1
Length=497
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 42/127 (33%), Positives = 69/127 (54%), Gaps = 19/127 (14%)
Query 24 FVRVTTKYWVRPENIVRAQCLIVRHLPFLVFGTSDKDLEASLLGPEAT--ANGHNFRGPN 81
F+R+T KYWV EN+V + I +HLP VF +++ P ++ N H+
Sbjct 165 FIRMTDKYWVHKENVVPVKFYITQHLPIYVFSKGNEE-----KSPYSSWDKNSHD----- 214
Query 82 LAPTQMVASVYFDSSSAYSYERRIRRFEGAQLLRFRWYGTNNDGPDEDIFIERKTHHESW 141
++SVYFD+ + Y R+++ ++ +R RWY + + PD +F+ERK H + W
Sbjct 215 ----TCISSVYFDNVAFDLYGERLKKLHHSEAIRIRWYTS--EVPDV-VFVERKKHEDGW 267
Query 142 SGMSSTK 148
+G SS K
Sbjct 268 TGESSKK 274
> YPL019c_1
Length=687
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 74/150 (49%), Gaps = 32/150 (21%)
Query 30 KYWVRPENIVRAQCLIVRHLPFLVFGTSDK-------DLEASL-LGPEA----------- 70
K+WV +NI+ + I+RHLP LV+ + +LE+ + + PEA
Sbjct 209 KFWVHDDNIMEVKARILRHLPALVYASVPNENDDFVDNLESDVRVQPEARLNIGSKSNSL 268
Query 71 TANGHNFRGPNLAPTQMV----------ASVYFDSSSAYSYERRIRRFEGAQLLRFRWYG 120
+++G++ + + ++ V ++YFD+ Y R+ + GA LR RW G
Sbjct 269 SSDGNSNQDVEIGKSKSVIFPQSYDPTITTLYFDNDFFDLYNNRLLKISGAPTLRLRWIG 328
Query 121 TNNDGPDEDIFIERKTHHE-SWSGMSSTKE 149
D P DIF+E++T E + +G SS +E
Sbjct 329 KLLDKP--DIFLEKRTFTENTETGNSSFEE 356
> YFL004w_1
Length=675
Score = 54.3 bits (129), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 57/122 (46%), Gaps = 19/122 (15%)
Query 30 KYWVRPENIVRAQCLIVRHLPFLVFGT--SDKDLEASLLGPEATANG------------- 74
K+WV +N++ + I+RHLP LV+ S+ D + + + N
Sbjct 207 KFWVHNDNLMEVKTRILRHLPVLVYANVPSENDDLVNRFESDISNNDEIVGSSSSTSSVE 266
Query 75 HNFRGPNLAPTQMVASVYFDSSSAYSYERRIRRFEGAQLLRFRWYGTNNDGPDEDIFIER 134
H + P ++ ++YFD+ Y ++ + A LR RW G +D P DIF+E+
Sbjct 267 HGLGARSFDP--LINTLYFDNEHFELYNDKLLKLNSAPTLRLRWTGQLSDKP--DIFLEK 322
Query 135 KT 136
KT
Sbjct 323 KT 324
> SPAC14C4.11_1
Length=606
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 53/116 (45%), Gaps = 25/116 (21%)
Query 24 FVRVTTKYWVRPENIVRAQCLIVRHLPFLVFGTS---DKDLE--ASLLGPEATANGHNFR 78
+ R T ++WV P+N++ + I+RHLP L + DKD + +L P +T
Sbjct 199 YRRRTFRFWVHPDNVMEVKTYIMRHLPVLYYSGKQGFDKDQNGVSGILDPISTC------ 252
Query 79 GPNLAPTQMVASVYFDSSSAYSYERRIRRFEGAQLLRFRWYGTNNDGPDEDIFIER 134
+Y D+S+ Y + + R E A LR WYG P DI +ER
Sbjct 253 ------------LYLDNSNFDLYSQNLERSEQAYSLRLHWYGKLT--PKTDIIVER 294
> CE07550
Length=574
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query 83 APTQMVASVYFDSSSAYSYERRIRRFEGAQLLRFR---WYGTNNDGPDEDIFIERKTHHE 139
AP +A + + +S R+RRFEG R R +G++ND PD + + +
Sbjct 44 APGAHIAPAHPEKTSGRDSRERVRRFEGNSPPRERDPDPFGSHNDAPDGNDYFGGDKDYR 103
Query 140 SWSGMSSTKE 149
S + SS +E
Sbjct 104 SGAAASSEEE 113
> Hs10863913
Length=973
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 23/95 (24%), Positives = 39/95 (41%), Gaps = 17/95 (17%)
Query 3 RFRSKLE---GRQMESEAWKPPDSFVRVTTKYWVRPENIVRAQCLIVRHLPFLVFGTSDK 59
RF S +E ++ E KP + T + W V +L + G+S
Sbjct 98 RFMSSIEVITSQEKEITIKKPNGETTKTTVRIWNET----------VSNLTLMALGSSAP 147
Query 60 DLEASLLGPEATANGHNFRGPNLAPTQMVASVYFD 94
++ S++ GHNF +L P+ +V S F+
Sbjct 148 EILLSVI----EVCGHNFTAGDLGPSTIVGSAAFN 178
> At2g04490
Length=1015
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 31/61 (50%), Gaps = 4/61 (6%)
Query 38 IVRAQCLIVRHLPFL--VFGTSDKDLEASLLGPEATANGHNFRGPNLAPTQMVASVYFDS 95
++R+Q ++ +H+P L ++ D+DL + P A NF + PTQ + ++
Sbjct 187 VIRSQIIMKKHIPELAGIYNLLDQDLSQRNITPVQNAVAFNFSA--MEPTQASVNATYNH 244
Query 96 S 96
S
Sbjct 245 S 245
> At1g53260
Length=358
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 55 GTSDKDLEASLLGPEATANGHNFRGPNLAPTQM 87
G ++ S GP + G N+RGP+L P M
Sbjct 279 GPPPPNMNQSYQGPPPSNMGQNYRGPSLPPPNM 311
> Hs10835031
Length=583
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query 2 GRFRSKLEGRQMESEAWKPPDSFVRVTTKYWVRPENIVRAQCL--IVRHL 49
G R+++EG Q+ E W SFV T+ W+ P + V + +VR++
Sbjct 118 GGRRTRVEGGQLGGEEWTRHGSFVNKPTRGWLHPNDKVMGPGVSYLVRYM 167
> Hs18584288
Length=238
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 21/38 (55%), Gaps = 6/38 (15%)
Query 45 IVRHLPFLVFGTSDKDLEASLLGPEATANGHNFRGPNL 82
++RHLP+LV G D+D + A GH+ PN+
Sbjct 175 LIRHLPYLVLGPGDRDYSGKF---QYRATGHH---PNI 206
> Hs6382079_1
Length=3065
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 31/76 (40%), Gaps = 4/76 (5%)
Query 67 GPEATANGHN----FRGPNLAPTQMVASVYFDSSSAYSYERRIRRFEGAQLLRFRWYGTN 122
GP++ +G+ F G L SVY+ S AY+ + +R + YG N
Sbjct 847 GPDSVPDGYQGSQTFHGAPLTVATTGPSVYYSQSPAYNSQYLLRPAANVTPTKGPVYGMN 906
Query 123 NDGPDEDIFIERKTHH 138
P + I+ + H
Sbjct 907 RLPPQQHIYAYPQQMH 922
Lambda K H
0.319 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40