bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
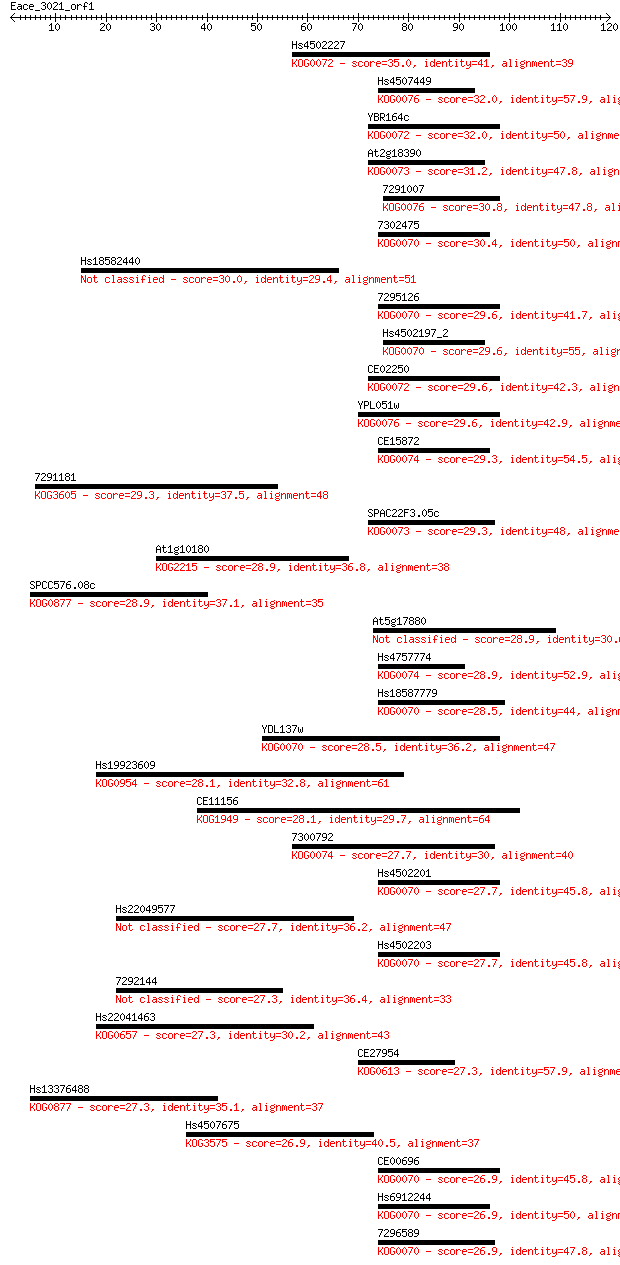
Query= Eace_3021_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
Hs4502227 35.0 0.034
Hs4507449 32.0 0.27
YBR164c 32.0 0.28
At2g18390 31.2 0.45
7291007 30.8 0.69
7302475 30.4 0.82
Hs18582440 30.0 0.98
7295126 29.6 1.4
Hs4502197_2 29.6 1.5
CE02250 29.6 1.5
YPL051w 29.6 1.5
CE15872 29.3 1.7
7291181 29.3 1.8
SPAC22F3.05c 29.3 1.8
At1g10180 28.9 2.3
SPCC576.08c 28.9 2.3
At5g17880 28.9 2.6
Hs4757774 28.9 2.7
Hs18587779 28.5 3.3
YDL137w 28.5 3.4
Hs19923609 28.1 3.7
CE11156 28.1 4.6
7300792 27.7 5.0
Hs4502201 27.7 5.1
Hs22049577 27.7 5.1
Hs4502203 27.7 5.2
7292144 27.3 7.2
Hs22041463 27.3 7.6
CE27954 27.3 7.7
Hs13376488 27.3 7.7
Hs4507675 26.9 7.9
CE00696 26.9 8.0
Hs6912244 26.9 8.3
7296589 26.9 9.9
> Hs4502227
Length=181
Score = 35.0 bits (79), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 4/43 (9%)
Query 57 SCSLCSSLSLPAFSG----VFACSAKTGEGIEEGISWLIRQLK 95
S + +SL LPA +F SA G G++E + WL+ LK
Sbjct 136 SSEMANSLGLPALKDRKWQIFKTSATKGTGLDEAMEWLVETLK 178
> Hs4507449
Length=201
Score = 32.0 bits (71), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 11/19 (57%), Positives = 16/19 (84%), Gaps = 0/19 (0%)
Query 74 ACSAKTGEGIEEGISWLIR 92
ACSA TG+G+ EGI W+++
Sbjct 167 ACSALTGKGVREGIEWMVK 185
> YBR164c
Length=183
Score = 32.0 bits (71), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 72 VFACSAKTGEGIEEGISWLIRQLKQQ 97
+ A SA GEGI EG+ WLI +K++
Sbjct 156 IVASSAIKGEGITEGLDWLIDVIKEE 181
> At2g18390
Length=185
Score = 31.2 bits (69), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 72 VFACSAKTGEGIEEGISWLIRQL 94
+ CSA TGEG+ EG WL++ +
Sbjct 155 IVGCSAYTGEGLLEGFDWLVQDI 177
> 7291007
Length=196
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 75 CSAKTGEGIEEGISWLIRQLKQQ 97
SA GEG++EGI WL+ +K+
Sbjct 164 VSALHGEGVDEGIKWLVEAIKRH 186
> 7302475
Length=202
Score = 30.4 bits (67), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLK 95
+ SA +GEG+ EG+ WLI+Q++
Sbjct 163 SSSAISGEGLGEGVQWLIQQMR 184
> Hs18582440
Length=222
Score = 30.0 bits (66), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 15 SGNTTSVTVASACGQGMVRLSPPPKRETLHVKYYFPSARIRLSCSLCSSLS 65
+G + +TVAS C QG+V++S +RE + + R+R + ++ ++L+
Sbjct 116 AGGASGITVASCCAQGVVKVSSQERREGTLALHTARNPRLRSAATVQATLA 166
> 7295126
Length=179
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLKQQ 97
AC A TGEG+ +G+ W+++++K +
Sbjct 156 ACCALTGEGLYQGLEWIVQRIKNK 179
> Hs4502197_2
Length=173
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 11/20 (55%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 75 CSAKTGEGIEEGISWLIRQL 94
C A++G G+ EG+ WL RQL
Sbjct 145 CDARSGMGLYEGLDWLSRQL 164
> CE02250
Length=180
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 72 VFACSAKTGEGIEEGISWLIRQLKQQ 97
+F SA GEG++ + WL QL+Q+
Sbjct 154 IFKTSASKGEGLDPAMDWLANQLQQK 179
> YPL051w
Length=198
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 70 SGVFACSAKTGEGIEEGISWLIRQLKQQ 97
S V SA TGEG+++ I W+I +L++
Sbjct 162 SRVLPISALTGEGVKDAIEWMIVRLERN 189
> CE15872
Length=184
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLK 95
ACSA EGI +GI+W+ LK
Sbjct 158 ACSALKNEGINDGITWVASNLK 179
> 7291181
Length=930
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 21/48 (43%), Gaps = 0/48 (0%)
Query 6 AGQAGCSIGSGNTTSVTVASACGQGMVRLSPPPKRETLHVKYYFPSAR 53
AG GSGN T+ A+A G G+ LS K T Y F R
Sbjct 20 AGSTANGTGSGNGTATGNATAAGSGLGPLSLHTKSSTFDYLYEFSETR 67
> SPAC22F3.05c
Length=186
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 72 VFACSAKTGEGIEEGISWLIRQLKQ 96
+F+ SA TG I++ ISWL LK+
Sbjct 154 IFSVSALTGLNIKDAISWLANDLKE 178
> At1g10180
Length=769
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 30 GMVRLSPPPKRETLHVKYYFPSARIRLSCSLCSSLSLP 67
G++RL P L +K+Y S R R+ L S L+ P
Sbjct 206 GLIRLGKGPSAHQLLLKFYATSLRRRIEAFLPSCLTCP 243
> SPCC576.08c
Length=253
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 5 KAGQAGCSIGSGNTTSVTVASACGQGMVRLSPPPK 39
+ G G ++G +T V V+ CG VRL P P+
Sbjct 139 RRGYWGTALGDPHTVPVKVSGKCGSVTVRLVPAPR 173
> At5g17880
Length=1197
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 11/36 (30%), Positives = 17/36 (47%), Gaps = 0/36 (0%)
Query 73 FACSAKTGEGIEEGISWLIRQLKQQDGAPCACDADQ 108
F+C GEG I+W + L +QD ++D
Sbjct 1021 FSCEQNNGEGSSSSITWKVGSLIEQDNQEETVESDH 1056
> Hs4757774
Length=182
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 9/17 (52%), Positives = 16/17 (94%), Gaps = 0/17 (0%)
Query 74 ACSAKTGEGIEEGISWL 90
+CSA TGEG+++G++W+
Sbjct 157 SCSALTGEGVQDGMNWV 173
> Hs18587779
Length=91
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLKQQD 98
A A TG G+ EG++WL Q + Q+
Sbjct 67 ATCATTGHGLYEGLNWLANQFQNQN 91
> YDL137w
Length=181
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 24/47 (51%), Gaps = 2/47 (4%)
Query 51 SARIRLSCSLCSSLSLPAFSGVFACSAKTGEGIEEGISWLIRQLKQQ 97
+A I L S + P F + + A +GEG+ EG+ WL LK Q
Sbjct 136 AAEITEKLGLHSIRNRPWF--IQSTCATSGEGLYEGLEWLSNNLKNQ 180
> Hs19923609
Length=509
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 26/61 (42%), Gaps = 4/61 (6%)
Query 18 TTSVTVASACGQGMVRLSPPPKRETLHVKYYFPSARIRLSCSLCSSLSLPAFSGVFACSA 77
T V V+ A V + P K E + + PS+R L CSLC+ F CS
Sbjct 275 TKWVHVSCALWIPEVSIGSPEKMEPITKVSHIPSSRWALVCSLCNE----KFGASIQCSV 330
Query 78 K 78
K
Sbjct 331 K 331
> CE11156
Length=1010
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 30/66 (45%), Gaps = 2/66 (3%)
Query 38 PKRETLHVKYYFPSARIRLSCSLCSSLSLPAFSGVFAC--SAKTGEGIEEGISWLIRQLK 95
P ++L YY P L S + LS G +++ + IE I+W R L+
Sbjct 374 PISKSLDEAYYRPRVNYMLGKSRLALLSYFRLLGPMKIIDASQASKLIEMFITWAYRYLR 433
Query 96 QQDGAP 101
++DG P
Sbjct 434 KKDGPP 439
> 7300792
Length=203
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 12/44 (27%), Positives = 24/44 (54%), Gaps = 4/44 (9%)
Query 57 SCSLCSSLSLPAFSG----VFACSAKTGEGIEEGISWLIRQLKQ 96
+ + +SL G + AC+A G G++EG+ W+ + +K+
Sbjct 160 AAEVAEKMSLVQLQGRTWEIKACTAVDGTGLKEGMDWVCKNMKK 203
> Hs4502201
Length=181
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLKQQ 97
A A +G+G+ EG+ WL QL+ Q
Sbjct 157 ATCATSGDGLYEGLDWLSNQLRNQ 180
> Hs22049577
Length=108
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 23/47 (48%), Gaps = 8/47 (17%)
Query 22 TVASACGQGMVRLSPPPKRETLHVKYYFPSARIRLSCSLCSSLSLPA 68
V++ G G V L P + YF S + R CS+ SL+LPA
Sbjct 57 NVSNLKGSGTVTLGP--------IVRYFGSWKARFGCSVSRSLNLPA 95
> Hs4502203
Length=181
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLKQQ 97
A A +G+G+ EG+ WL QLK +
Sbjct 157 ATCATSGDGLYEGLDWLANQLKNK 180
> 7292144
Length=1205
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 22 TVASACGQGMVRLSPPPKRETLHVKYYFPSARI 54
TV + CG + +SP +R+T V +Y P +I
Sbjct 270 TVLAICGTTLDAVSPTSQRDTNQVCFYSPLGKI 302
> Hs22041463
Length=277
Score = 27.3 bits (59), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 13/46 (28%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 18 TTSVTVASACGQGMVRLSPPPKRETLHVKYYFPSARI---RLSCSL 60
T+S+ V++ + + ++ P P R+ V ++ P+A + L+CSL
Sbjct 161 TSSLHVSTGAAKAVGKVIPKPNRKLSGVAFHVPTANVSVMDLTCSL 206
> CE27954
Length=3484
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 70 SGVFACSAKTGEGIEEGIS 88
SGVF C K+ EG EE +S
Sbjct 3048 SGVFTCKVKSSEGDEEEVS 3066
> Hs13376488
Length=148
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 16/37 (43%), Gaps = 0/37 (0%)
Query 5 KAGQAGCSIGSGNTTSVTVASACGQGMVRLSPPPKRE 41
+ G G IG TT V C G+V L P P+
Sbjct 2 RRGYWGNKIGKSYTTPCMVTGCCSSGLVHLIPAPRNN 38
> Hs4507675
Length=615
Score = 26.9 bits (58), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 36 PPPKRETLHVKYYFPSARIRLSCSLCSSLSLPAFSGV 72
P P E L+V Y SA L S+ + S+PAF G+
Sbjct 393 PSPMPEYLNVHYICESASRLLFLSMHRARSIPAFQGL 429
> CE00696
Length=181
Score = 26.9 bits (58), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLKQQ 97
A A +G+G+ EG+ WL QLK +
Sbjct 157 ATCATSGDGLYEGLDWLSNQLKNR 180
> Hs6912244
Length=179
Score = 26.9 bits (58), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLK 95
AC A TGEG+ +G+ W++ +LK
Sbjct 156 ACCALTGEGLCQGLEWMMSRLK 177
> 7296589
Length=182
Score = 26.9 bits (58), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 74 ACSAKTGEGIEEGISWLIRQLKQ 96
A A +G+G+ EG+ WL QLK
Sbjct 157 ATCATSGDGLYEGLDWLSNQLKN 179
Lambda K H
0.320 0.133 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40