bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3062_orf1
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
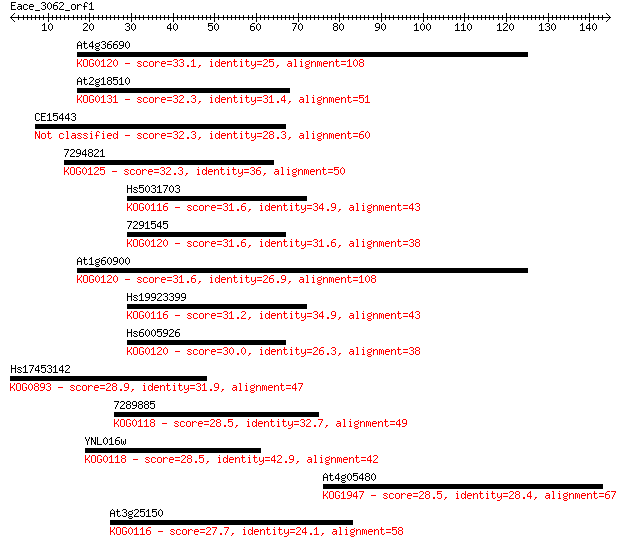
At4g36690 33.1 0.17
At2g18510 32.3 0.30
CE15443 32.3 0.31
7294821 32.3 0.31
Hs5031703 31.6 0.52
7291545 31.6 0.55
At1g60900 31.6 0.58
Hs19923399 31.2 0.65
Hs6005926 30.0 1.5
Hs17453142 28.9 3.4
7289885 28.5 3.9
YNL016w 28.5 4.3
At4g05480 28.5 4.6
At3g25150 27.7 7.2
> At4g36690
Length=573
Score = 33.1 bits (74), Expect = 0.17, Method: Composition-based stats.
Identities = 27/112 (24%), Positives = 55/112 (49%), Gaps = 10/112 (8%)
Query 17 ILHVWFCKERGSNYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWRRDAADLNFAD 76
+++V+ E+ + FVE+ S E+A +S+ I+++G +++ R +++ A A
Sbjct 277 VVNVYINHEK--KFAFVEMRSVEEASNAMSLDGIIFEGAPVKVRRPSDYNPSLA----AT 330
Query 77 VAGSANAPLLESIAAAVSLAETTG----DRRGLMLLLARLTETQRNVLVDFF 124
+ S +P L A ++ + G DR + L TE+Q L++ F
Sbjct 331 LGPSQPSPHLNLAAVGLTPGASGGLEGPDRIFVGGLPYYFTESQVRELLESF 382
> At2g18510
Length=363
Score = 32.3 bits (72), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 33/56 (58%), Gaps = 5/56 (8%)
Query 17 ILHVWFCKER----GSNYGFVELSSAEDAQRLLSI-HPIVWKGQQLRINRHTEWRR 67
+++V+ K+R NYGF+E S EDA + + + I G+ +R+N+ ++ ++
Sbjct 51 VVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAIKVLNMIKLHGKPIRVNKASQDKK 106
> CE15443
Length=347
Score = 32.3 bits (72), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query 7 QKKETSIYSCILHVWFCKERGSNYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWR 66
+ K+ I CI ++F + + F++ + ED ++L+ IH +K Q IN H + R
Sbjct 122 KAKQLIILICI--IYFPSYLQAIFVFIDNNKPEDVRQLIQIHHSQYKLQDFVINGHADLR 179
> 7294821
Length=287
Score = 32.3 bits (72), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 30/53 (56%), Gaps = 3/53 (5%)
Query 14 YSCILHVWFC-KERGS-NYGFVELSSAEDAQRLLS-IHPIVWKGQQLRINRHT 63
+ IL V ERGS +GFV +++ DA+R +H V +G+++ +N T
Sbjct 220 FGTILDVEIIFNERGSKGFGFVTFANSNDAERARERLHGTVVEGRKIEVNNAT 272
> Hs5031703
Length=466
Score = 31.6 bits (70), Expect = 0.52, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 27/43 (62%), Gaps = 1/43 (2%)
Query 29 NYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWRRDAAD 71
N+GFV +E Q++LS PI+++G ++R+N + R A +
Sbjct 379 NFGFVVFDDSEPVQKVLSNRPIMFRG-EVRLNVEEKKTRAARE 420
> 7291545
Length=449
Score = 31.6 bits (70), Expect = 0.55, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 29 NYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWR 66
N+ F+E S ++A + L+ +V++GQ L+I R +++
Sbjct 163 NFAFLEFRSIDEASQALNFDGMVFRGQTLKIRRPHDYQ 200
> At1g60900
Length=568
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 29/112 (25%), Positives = 52/112 (46%), Gaps = 10/112 (8%)
Query 17 ILHVWFCKERGSNYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWRRDAADLNFAD 76
+++V+ E+ + FVE+ S E+A +++ I+ +G +++ R T++ A A
Sbjct 272 VVNVYINHEK--KFAFVEMRSVEEASNAMALDGIILEGVPVKVRRPTDYNPSLA----AT 325
Query 77 VAGSANAPLLESIAAAVSLAETTG----DRRGLMLLLARLTETQRNVLVDFF 124
+ S P L A +S T G DR + L TE Q L++ F
Sbjct 326 LGPSQPNPNLNLGAVGLSSGSTGGLEGPDRIFVGGLPYYFTEVQIRELLESF 377
> Hs19923399
Length=482
Score = 31.2 bits (69), Expect = 0.65, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 26/43 (60%), Gaps = 1/43 (2%)
Query 29 NYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWRRDAAD 71
N+GFV +E QR+L PI+++G ++R+N + R A +
Sbjct 373 NFGFVVFDDSEPVQRILIAKPIMFRG-EVRLNVEEKKTRAARE 414
> Hs6005926
Length=475
Score = 30.0 bits (66), Expect = 1.5, Method: Composition-based stats.
Identities = 10/38 (26%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 29 NYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWR 66
N+ F+E S ++ + ++ I+++GQ L+I R +++
Sbjct 196 NFAFLEFRSVDETTQAMAFDGIIFQGQSLKIRRPHDYQ 233
> Hs17453142
Length=346
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 25/54 (46%), Gaps = 7/54 (12%)
Query 1 SSNCLQQKKETSIYSC-------ILHVWFCKERGSNYGFVELSSAEDAQRLLSI 47
S C++Q K TS++ C + WF G GF + ++DA + S+
Sbjct 222 SHGCVRQGKRTSVFCCGCGGRFQCMQCWFTSSAGRVPGFSPPAFSDDAWEVASL 275
> 7289885
Length=507
Score = 28.5 bits (62), Expect = 3.9, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Query 26 RGSNYGFVELSSAEDAQRLLSIHPIVWKGQQLRI---NRHTEWRRDAADLNF 74
R +G+VEL + ED +LS+ KG+++RI N + + R ++ F
Sbjct 314 RSRGFGYVELENREDLIHVLSLPDPSIKGRRIRIELSNENDQQSRQKSNRRF 365
> YNL016w
Length=453
Score = 28.5 bits (62), Expect = 4.3, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 3/45 (6%)
Query 19 HVWFCKERGSN--YGFVELSSAEDAQRLL-SIHPIVWKGQQLRIN 60
HV + + GS+ YGFV +S +DAQ + S+ G+ LRIN
Sbjct 192 HVMWDMQTGSSRGYGFVSFTSQDDAQNAMDSMQGQDLNGRPLRIN 236
> At4g05480
Length=322
Score = 28.5 bits (62), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 30/67 (44%), Gaps = 0/67 (0%)
Query 76 DVAGSANAPLLESIAAAVSLAETTGDRRGLMLLLARLTETQRNVLVDFFTKCPSALTGSS 135
D++ PLLE A+++AE+ R L L+ LT T N ++D L
Sbjct 200 DISQVGYIPLLECDDDALAIAESMPKLRHLQLMGNGLTNTGLNAILDGCPHLEEHLDVRK 259
Query 136 CLPVELL 142
C + L+
Sbjct 260 CFNINLV 266
> At3g25150
Length=486
Score = 27.7 bits (60), Expect = 7.2, Method: Composition-based stats.
Identities = 14/58 (24%), Positives = 23/58 (39%), Gaps = 0/58 (0%)
Query 25 ERGSNYGFVELSSAEDAQRLLSIHPIVWKGQQLRINRHTEWRRDAADLNFADVAGSAN 82
++G +GFVE SA Q + P++ G ++ + R F G N
Sbjct 348 QKGFCFGFVEFESASSMQSAIEASPVMLNGHKVVVEEKRSTARGKGRSTFGVNTGYRN 405
Lambda K H
0.320 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1675978996
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40