bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3080_orf2
Length=67
Score E
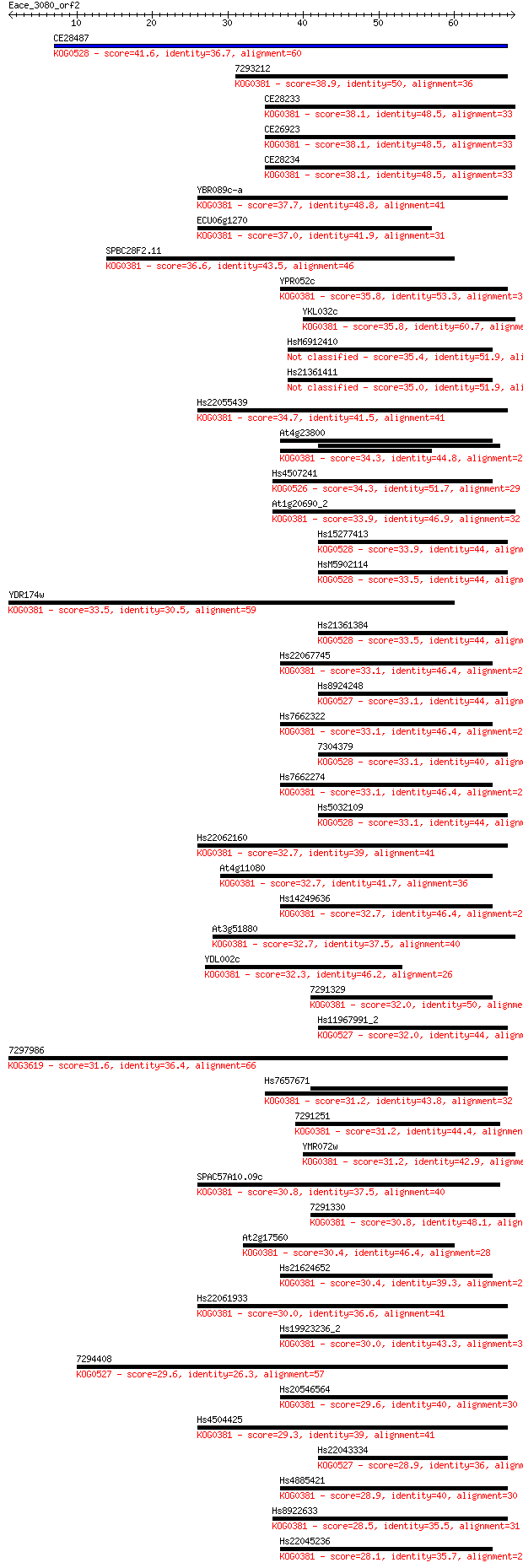
Sequences producing significant alignments: (Bits) Value
CE28487 41.6 3e-04
7293212 38.9 0.002
CE28233 38.1 0.004
CE26923 38.1 0.004
CE28234 38.1 0.004
YBR089c-a 37.7 0.005
ECU06g1270 37.0 0.010
SPBC28F2.11 36.6 0.012
YPR052c 35.8 0.018
YKL032c 35.8 0.019
HsM6912410 35.4 0.029
Hs21361411 35.0 0.029
Hs22055439 34.7 0.043
At4g23800 34.3 0.054
Hs4507241 34.3 0.063
At1g20690_2 33.9 0.073
Hs15277413 33.9 0.082
HsM5902114 33.5 0.089
YDR174w 33.5 0.092
Hs21361384 33.5 0.100
Hs22067745 33.1 0.11
Hs8924248 33.1 0.13
Hs7662322 33.1 0.13
7304379 33.1 0.13
Hs7662274 33.1 0.14
Hs5032109 33.1 0.14
Hs22062160 32.7 0.16
At4g11080 32.7 0.16
Hs14249636 32.7 0.17
At3g51880 32.7 0.17
YDL002c 32.3 0.21
7291329 32.0 0.25
Hs11967991_2 32.0 0.28
7297986 31.6 0.38
Hs7657671 31.2 0.43
7291251 31.2 0.43
YMR072w 31.2 0.52
SPAC57A10.09c 30.8 0.62
7291330 30.8 0.65
At2g17560 30.4 0.80
Hs21624652 30.4 0.88
Hs22061933 30.0 0.98
Hs19923236_2 30.0 1.2
7294408 29.6 1.3
Hs20546564 29.6 1.5
Hs4504425 29.3 1.6
Hs22043334 28.9 2.1
Hs4885421 28.9 2.5
Hs8922633 28.5 2.8
Hs22045236 28.1 3.8
> CE28487
Length=470
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 39/60 (65%), Gaps = 5/60 (8%)
Query 7 SSSSISSSSSSSSSSNVAAPRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
+SS S+ ++S +S VA PR +K P KRP++A+M++ARD+R ++L+ P++
Sbjct 300 NSSGKSTPGNTSVTSEVATPRPQAKS-----PNHIKRPMNAFMVWARDERRKILKAYPDM 354
> 7293212
Length=328
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 31 KRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
KR KDP APKR LSA+ F D+R +V PE
Sbjct 196 KRKQIKDPNAPKRSLSAFFWFCNDERNKVKALNPEF 231
> CE28233
Length=234
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 35 QKDPAAPKRPLSAYMLFARDKRVEVLQQQPELK 67
+KDP APKR LSA+ +++DKR E+ P+ K
Sbjct 128 KKDPHAPKRALSAFFFYSQDKRPEIQAGHPDWK 160
> CE26923
Length=235
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 35 QKDPAAPKRPLSAYMLFARDKRVEVLQQQPELK 67
+KDP APKR LSA+ +++DKR E+ P+ K
Sbjct 129 KKDPHAPKRALSAFFFYSQDKRPEIQAGHPDWK 161
> CE28234
Length=233
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 35 QKDPAAPKRPLSAYMLFARDKRVEVLQQQPELK 67
+KDP APKR LSA+ +++DKR E+ P+ K
Sbjct 127 KKDPHAPKRALSAFFFYSQDKRPEIQAGHPDWK 159
> YBR089c-a
Length=99
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 26 PRTSSKR--HHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
P+ KR +KDP APKR LSAYM FA + R V + P++
Sbjct 10 PKEPKKRTTRRKKDPNAPKRRLSAYMFFANENRDIVRSENPDV 52
> ECU06g1270
Length=201
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 26 PRTSSKRHHQKDPAAPKRPLSAYMLFARDKR 56
PR+ + +KDP APK+P+S Y +F +++R
Sbjct 29 PRSKTAVKRRKDPNAPKKPMSGYFIFGQEQR 59
> SPBC28F2.11
Length=310
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 30/46 (65%), Gaps = 1/46 (2%)
Query 14 SSSSSSSSNVAAPRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEV 59
++S+ +S A T +KR +DPA PKRP SAY LF +++R E+
Sbjct 91 ATSAVDTSEAVASMTPNKRKA-RDPAQPKRPPSAYNLFQKNQRSEI 135
> YPR052c
Length=93
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
DP APKR LSAYM FA + R V + P++
Sbjct 17 DPNAPKRALSAYMFFANENRDIVRSENPDI 46
> YKL032c
Length=597
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 17/28 (60%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 40 APKRPLSAYMLFARDKRVEVLQQQPELK 67
PKRP SAY LF+ R E+LQQ PE K
Sbjct 360 GPKRPSSAYFLFSMSIRNELLQQFPEAK 387
> HsM6912410
Length=514
Score = 35.4 bits (80), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 38 PAAPKRPLSAYMLFARDKRVEVLQQQP 64
P KRP++A+MLFA+ RVE Q P
Sbjct 431 PNKCKRPMNAFMLFAKKYRVEYTQMYP 457
> Hs21361411
Length=514
Score = 35.0 bits (79), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 38 PAAPKRPLSAYMLFARDKRVEVLQQQP 64
P KRP++A+MLFA+ RVE Q P
Sbjct 431 PNKCKRPMNAFMLFAKKYRVEYTQMYP 457
> Hs22055439
Length=168
Score = 34.7 bits (78), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 26 PRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
P+ +K+ KDP APKRP SA+ LF + R ++ + P L
Sbjct 49 PKGETKKKF-KDPNAPKRPPSAFFLFCSEYRPKIKGEHPGL 88
> At4g23800
Length=456
Score = 34.3 bits (77), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQP 64
DP PK+P S+Y LF++D+R ++ +++P
Sbjct 375 DPNKPKKPASSYFLFSKDERKKLTEERP 402
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPE 65
KRP S+Y+L+ +D+ EV ++ PE
Sbjct 139 KRPSSSYVLWCKDQWTEVKKENPE 162
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 9/20 (45%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 37 DPAAPKRPLSAYMLFARDKR 56
DP PK P+SA++++A ++R
Sbjct 251 DPLKPKHPVSAFLVYANERR 270
> Hs4507241
Length=709
Score = 34.3 bits (77), Expect = 0.063, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 36 KDPAAPKRPLSAYMLFARDKRVEVLQQQP 64
KDP APKRP+SAYML+ R ++ P
Sbjct 542 KDPNAPKRPMSAYMLWLNASREKIKSDHP 570
> At1g20690_2
Length=249
Score = 33.9 bits (76), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 36 KDPAAPKRPLSAYMLFARDKRVEVLQQQPELK 67
KDP PKRP SA+ +F D RV ++ P+ K
Sbjct 138 KDPNKPKRPSSAFFVFMEDFRVTYKEEHPKNK 169
> Hs15277413
Length=808
Score = 33.9 bits (76), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 22/25 (88%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
KRP++A+M++A+D+R ++LQ P++
Sbjct 602 KRPMNAFMVWAKDERRKILQAFPDM 626
> HsM5902114
Length=347
Score = 33.5 bits (75), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 22/25 (88%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
KRP++A+M++A+D+R ++LQ P++
Sbjct 141 KRPMNAFMVWAKDERRKILQAFPDM 165
> YDR174w
Length=246
Score = 33.5 bits (75), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 1 HLSGRRSSSSISSSSSSSSSSNVAAPRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEV 59
HL G SS + + + +AAP + +R ++DP APK+PL+ + ++ R E+
Sbjct 68 HLHG--ISSELVNPIDDDKDAIIAAPVKAVRRKIERDPNAPKKPLTVFFAYSAYVRQEL 124
> Hs21361384
Length=763
Score = 33.5 bits (75), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 22/25 (88%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
KRP++A+M++A+D+R ++LQ P++
Sbjct 557 KRPMNAFMVWAKDERRKILQAFPDM 581
> Hs22067745
Length=576
Score = 33.1 bits (74), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQP 64
DP P++P+SAY LF RD + + Q P
Sbjct 251 DPNEPQKPVSAYALFFRDTQAAIKGQNP 278
> Hs8924248
Length=384
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 21/25 (84%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
+RP++A+M++A+D+R + QQ P+L
Sbjct 86 RRPMNAFMVWAKDERKRLAQQNPDL 110
> Hs7662322
Length=526
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQP 64
DP P++P+SAY LF RD + + Q P
Sbjct 257 DPNEPQKPVSAYALFFRDTQAAIKGQNP 284
> 7304379
Length=580
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 10/25 (40%), Positives = 22/25 (88%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
KRP++A+M++A+D+R ++L+ P++
Sbjct 386 KRPMNAFMVWAKDERRKILKACPDM 410
> Hs7662274
Length=621
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQP 64
DP P++P+SAY LF RD + + Q P
Sbjct 219 DPNEPQKPVSAYALFFRDTQAAIKGQNP 246
> Hs5032109
Length=622
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 22/25 (88%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
KRP++A+M++A+D+R ++LQ P++
Sbjct 425 KRPMNAFMVWAKDERRKILQAFPDM 449
> Hs22062160
Length=187
Score = 32.7 bits (73), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Query 26 PRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
P+ +K+ KDP APKRP SA+ LF + ++ + P L
Sbjct 57 PKGETKKKF-KDPNAPKRPPSAFFLFCSEYHPKIKGEHPGL 96
> At4g11080
Length=446
Score = 32.7 bits (73), Expect = 0.16, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 29 SSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQP 64
+ K++ DP PK+P S+Y LF +D R VL++ P
Sbjct 360 NKKKNENVDPNKPKKPTSSYFLFCKDARKSVLEEHP 395
> Hs14249636
Length=464
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQP 64
DP P++P+SAY LF RD + + Q P
Sbjct 200 DPNEPQKPVSAYALFFRDTQAAIKGQNP 227
> At3g51880
Length=178
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 28 TSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPELK 67
T ++ +KDP PKR SA+ +F D RV ++ P +K
Sbjct 40 TRKEKKAKKDPNKPKRAPSAFFVFLEDFRVTFKKENPNVK 79
> YDL002c
Length=203
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 20/28 (71%), Gaps = 2/28 (7%)
Query 27 RTSSKRHH--QKDPAAPKRPLSAYMLFA 52
++ +KRH ++DP PKRP +AY+L+
Sbjct 78 KSKTKRHKVKERDPNMPKRPTNAYLLYC 105
> 7291329
Length=111
Score = 32.0 bits (71), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 41 PKRPLSAYMLFARDKRVEVLQQQP 64
PKRPLSAYML+ + R ++ + P
Sbjct 6 PKRPLSAYMLWLNETREQIKKDNP 29
> Hs11967991_2
Length=371
Score = 32.0 bits (71), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 21/25 (84%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
+RP++A+M++A+D+R + QQ P+L
Sbjct 26 RRPMNAFMVWAKDERKRLAQQNPDL 50
> 7297986
Length=856
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 35/71 (49%), Gaps = 15/71 (21%)
Query 1 HLSGRRSSSSISSSSSSSSSSNVAAPRTSSKRHHQKDPAAPKRPLSAYMLFA-----RDK 55
H+SGR + SS ++ N+ P T S QKDP K P+S Y+L A DK
Sbjct 347 HVSGR------TLSSLNAEEYNIY-PGTESA---QKDPVLQKHPMSTYLLTAIPSLDSDK 396
Query 56 RVEVLQQQPEL 66
+ +++ P L
Sbjct 397 TISIVEGVPNL 407
> Hs7657671
Length=764
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 41 PKRPLSAYMLFARDKRVEVLQQQPEL 66
PKRP+SA +F+ +KR ++ +++PEL
Sbjct 407 PKRPVSAMFIFSEEKRRQLQEERPEL 432
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 35 QKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
+K P PK+PL+ Y F +KR + + PE+
Sbjct 106 KKHPDFPKKPLTPYFRFFMEKRAKYAKLHPEM 137
> 7291251
Length=376
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 39 AAPKRPLSAYMLFARDKRVEVLQQQPE 65
APK PL+ Y+ F D+R E+ ++QP+
Sbjct 74 GAPKMPLNGYVRFMNDRREELRREQPQ 100
> YMR072w
Length=183
Score = 31.2 bits (69), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 40 APKRPLSAYMLFARDKRVEVLQQQPELK 67
PKRP SAY L+ +D R + +++ P L+
Sbjct 42 GPKRPTSAYFLYLQDHRSQFVKENPTLR 69
> SPAC57A10.09c
Length=108
Score = 30.8 bits (68), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 26 PRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPE 65
PR ++K +KDP PKR +SA+M F+ + R ++ P+
Sbjct 2 PR-AAKSSRKKDPNTPKRNMSAFMFFSIENREKMKTDNPD 40
> 7291330
Length=112
Score = 30.8 bits (68), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 41 PKRPLSAYMLFARDKRVEVLQQQPELK 67
PKRPLSAYML+ R + ++ P +K
Sbjct 5 PKRPLSAYMLWLNSARESIKRENPGIK 31
> At2g17560
Length=138
Score = 30.4 bits (67), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 32 RHHQKDPAAPKRPLSAYMLFARDKRVEV 59
+ +KDP PKRP SA+ +F D R E
Sbjct 26 KKTKKDPNQPKRPPSAFFVFLEDFRKEF 53
> Hs21624652
Length=186
Score = 30.4 bits (67), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQP 64
DP AP+RP S+++LF +D ++ ++ P
Sbjct 89 DPQAPRRPPSSFLLFCQDHYAQLKRENP 116
> Hs22061933
Length=411
Score = 30.0 bits (66), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 26 PRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
P+ +K+ KDP APKRP SA+ L+ + ++ ++P L
Sbjct 285 PKGETKKKF-KDPNAPKRPPSAFFLYFSEYGPKIKGERPGL 324
> Hs19923236_2
Length=181
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
DP APKRP A+ LF + R ++ + P L
Sbjct 67 DPNAPKRPPLAFFLFCSEYRPKIKGEHPGL 96
> 7294408
Length=217
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 10 SISSSSSSSSSSNVAAPRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
++S + S+ SS + S D KRP++A+M+++R +R ++ Q P++
Sbjct 151 ALSHQTHSNGSSQLGGGSASGAGSVAGDEEHIKRPMNAFMVWSRLQRRKIAQDNPKM 207
> Hs20546564
Length=173
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
DP+APKRP S + LF+ + R ++ P +
Sbjct 84 DPSAPKRPPSGFFLFSSEIRPKIKSTNPGI 113
> Hs4504425
Length=215
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 26 PRTSSKRHHQKDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
P + KDP APKRP SA+ LF + R ++ + P L
Sbjct 80 PPKGETKKKFKDPNAPKRPPSAFFLFCSEYRPKIKGEHPGL 120
> Hs22043334
Length=317
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 9/25 (36%), Positives = 21/25 (84%), Gaps = 0/25 (0%)
Query 42 KRPLSAYMLFARDKRVEVLQQQPEL 66
KRP++A+M+++R +R ++ Q+ P++
Sbjct 42 KRPMNAFMVWSRGQRRKMAQENPKM 66
> Hs4885421
Length=200
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
DP APKRP S + LF + R ++ P +
Sbjct 89 DPNAPKRPPSGFFLFCSEFRPKIKSTNPGI 118
> Hs8922633
Length=347
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 36 KDPAAPKRPLSAYMLFARDKRVEVLQQQPEL 66
+D APK PL+ Y+ F ++R ++ ++PE+
Sbjct 98 RDSNAPKSPLTGYVRFMNERREQLRAKRPEV 128
> Hs22045236
Length=112
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 10/28 (35%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 37 DPAAPKRPLSAYMLFARDKRVEVLQQQP 64
DP P+RP S+++LF +D ++ ++ P
Sbjct 15 DPQEPRRPPSSFLLFCQDHYAQLKRENP 42
Lambda K H
0.304 0.114 0.296
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160559522
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40