bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3101_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
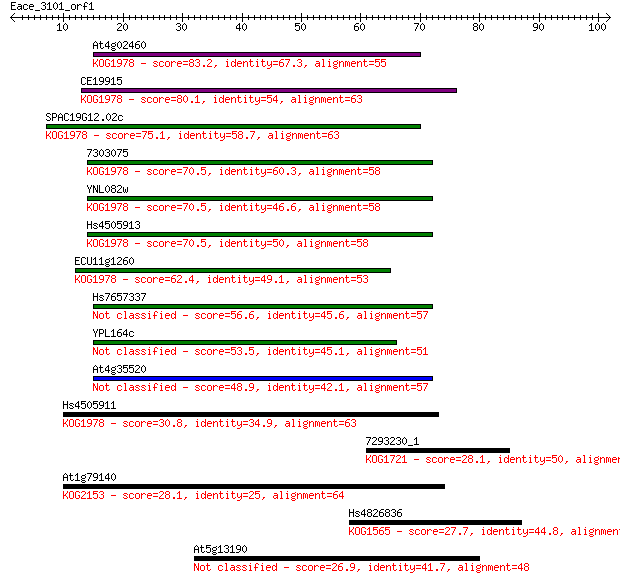
At4g02460 83.2 1e-16
CE19915 80.1 9e-16
SPAC19G12.02c 75.1 3e-14
7303075 70.5 7e-13
YNL082w 70.5 8e-13
Hs4505913 70.5 8e-13
ECU11g1260 62.4 2e-10
Hs7657337 56.6 1e-08
YPL164c 53.5 9e-08
At4g35520 48.9 3e-06
Hs4505911 30.8 0.68
7293230_1 28.1 4.2
At1g79140 28.1 4.6
Hs4826836 27.7 5.8
At5g13190 26.9 8.7
> At4g02460
Length=779
Score = 83.2 bits (204), Expect = 1e-16, Method: Composition-based stats.
Identities = 37/55 (67%), Positives = 44/55 (80%), Gaps = 0/55 (0%)
Query 15 PSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTMRHLV 69
PSRV +LASRACRS+VMIGDPL + Q I+ +A L+ P+NCPHGRPTMRHLV
Sbjct 698 PSRVRAMLASRACRSSVMIGDPLRKNEMQKIVEHLADLESPWNCPHGRPTMRHLV 752
> CE19915
Length=805
Score = 80.1 bits (196), Expect = 9e-16, Method: Composition-based stats.
Identities = 34/63 (53%), Positives = 45/63 (71%), Gaps = 0/63 (0%)
Query 13 YRPSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTMRHLVDIP 72
YRP R+ I AS+ACR +VMIG PL+ ++ I+R +A L P+NCPHGRPT+RHL +P
Sbjct 740 YRPVRIRKIFASKACRKSVMIGKPLNQREMTQIIRHLAKLDQPWNCPHGRPTIRHLASLP 799
Query 73 TAA 75
A
Sbjct 800 DRA 802
> SPAC19G12.02c
Length=794
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 37/69 (53%), Positives = 49/69 (71%), Gaps = 6/69 (8%)
Query 7 IITVLQYRP------SRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPH 60
II+VL P SR+ +LAS+ACRS+VMIG L++ + TI+R +A L P+NCPH
Sbjct 721 IISVLSEHPQIDPFSSRLERMLASKACRSSVMIGRALTISEMNTIVRHLAELSKPWNCPH 780
Query 61 GRPTMRHLV 69
GRPTMRHL+
Sbjct 781 GRPTMRHLL 789
> 7303075
Length=899
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 35/62 (56%), Positives = 44/62 (70%), Gaps = 7/62 (11%)
Query 14 RPSRVWNILASRACRSAVMIGDPLSLQKQQTILRR----MALLQLPFNCPHGRPTMRHLV 69
RPSRV + ASRACR +VMIG LS + T +RR M ++ P+NCPHGRPTMRHL+
Sbjct 825 RPSRVRAMFASRACRKSVMIGTALS---RNTTMRRLITQMGEIEQPWNCPHGRPTMRHLI 881
Query 70 DI 71
+I
Sbjct 882 NI 883
> YNL082w
Length=904
Score = 70.5 bits (171), Expect = 8e-13, Method: Composition-based stats.
Identities = 27/58 (46%), Positives = 43/58 (74%), Gaps = 0/58 (0%)
Query 14 RPSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTMRHLVDI 71
R S++ ++ A RACRS++MIG PL+ + ++ ++ L P+NCPHGRPTMRHL+++
Sbjct 835 RCSKIRSMFAMRACRSSIMIGKPLNKKTMTRVVHNLSELDKPWNCPHGRPTMRHLMEL 892
> Hs4505913
Length=862
Score = 70.5 bits (171), Expect = 8e-13, Method: Composition-based stats.
Identities = 29/58 (50%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 14 RPSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTMRHLVDI 71
RPSRV + ASRACR +VMIG L+ + + ++ M + P+NCPHGRPTMRH+ ++
Sbjct 799 RPSRVKQMFASRACRKSVMIGTALNTSEMKKLITHMGEMDHPWNCPHGRPTMRHIANL 856
> ECU11g1260
Length=630
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 26/53 (49%), Positives = 41/53 (77%), Gaps = 2/53 (3%)
Query 12 QYRPSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPT 64
++R +R +I+AS+ACR++VMIGD LS+ + I++ +A L+ P+ CPHGRPT
Sbjct 571 EFRKAR--SIIASKACRTSVMIGDVLSMADMKRIVKSLASLERPWKCPHGRPT 621
> Hs7657337
Length=1429
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 35/57 (61%), Gaps = 0/57 (0%)
Query 15 PSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTMRHLVDI 71
P V +LAS+AC A+ D LSLQ+ ++ ++ QLPF C HGRP+M L DI
Sbjct 1326 PLTVQKVLASQACHGAIKFNDGLSLQESCRLIEALSSCQLPFQCAHGRPSMLPLADI 1382
> YPL164c
Length=715
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 23/51 (45%), Positives = 33/51 (64%), Gaps = 0/51 (0%)
Query 15 PSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTM 65
P+ IL S+ACRSAVM GD L+ Q+ ++ +++ PF C HGRP+M
Sbjct 658 PTVFHEILNSKACRSAVMFGDELTRQECIILISKLSRCHNPFECAHGRPSM 708
> At4g35520
Length=1151
Score = 48.9 bits (115), Expect = 3e-06, Method: Composition-based stats.
Identities = 24/57 (42%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 15 PSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTMRHLVDI 71
P V +L S+ACR A+M GD L + I+ + L F C HGRPT LVD+
Sbjct 1057 PPSVLRVLNSKACRGAIMFGDSLLPSECSLIIDGLKQTSLCFQCAHGRPTTVPLVDL 1113
> Hs4505911
Length=932
Score = 30.8 bits (68), Expect = 0.68, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 31/68 (45%), Gaps = 8/68 (11%)
Query 10 VLQYRPSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPF-----NCPHGRPT 64
V + RP +V + L A R + + LS + Q I+ RM + F C HGRP
Sbjct 865 VYECRPRKVISYLEGEAVRLSRQLPMYLSKEDIQDIIYRM---KHQFGNEIKECVHGRPF 921
Query 65 MRHLVDIP 72
HL +P
Sbjct 922 FHHLTYLP 929
> 7293230_1
Length=506
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 61 GRPTMRHLVDIPTAAAVGAAAAAP 84
G+PT L D+ T AA G+ A+AP
Sbjct 55 GKPTSSDLADLSTLAAGGSTASAP 78
> At1g79140
Length=311
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query 10 VLQYRPSRVWNILASRACRSAVMIGDPLSLQKQQTILRRMALLQLPFNCPHGRPTMRHLV 69
+L+YRP R ++ + + + + +QK ++R+A L F C +M LV
Sbjct 68 ILEYRPGRDSGVILAESLKIMLCDDRHQDMQKAAAFVKRLATFALCFGCAE---SMSALV 124
Query 70 DIPT 73
+ T
Sbjct 125 TLKT 128
> Hs4826836
Length=707
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 16/29 (55%), Gaps = 1/29 (3%)
Query 58 CPHGRPTMRHLVDIPTAAAVGAAAAAPTA 86
CP G PT+ H + PTA G +A PT
Sbjct 468 CPTGPPTV-HPSERPTAGPTGPPSAGPTG 495
> At5g13190
Length=146
Score = 26.9 bits (58), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 24/49 (48%), Gaps = 4/49 (8%)
Query 32 MIGDPLSLQKQQTILRRMALLQLPFNCPH-GRPTMRHLVDIPTAAAVGA 79
+ GDPL QQTI R PFNC + G + +L P AAV A
Sbjct 30 IYGDPLGAPIQQTIYRDTP---APFNCLYCGNTGLTNLRSKPGVAAVVA 75
Lambda K H
0.323 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40