bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
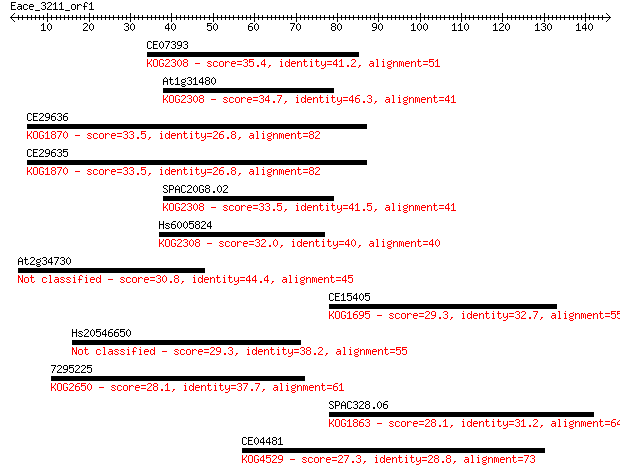
Query= Eace_3211_orf1
Length=145
Score E
Sequences producing significant alignments: (Bits) Value
CE07393 35.4 0.040
At1g31480 34.7 0.061
CE29636 33.5 0.15
CE29635 33.5 0.15
SPAC20G8.02 33.5 0.16
Hs6005824 32.0 0.41
At2g34730 30.8 0.86
CE15405 29.3 2.7
Hs20546650 29.3 2.8
7295225 28.1 5.4
SPAC328.06 28.1 6.7
CE04481 27.3 9.2
> CE07393
Length=765
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 28/51 (54%), Gaps = 3/51 (5%)
Query 34 QMPRRYDFQLQEDTAEHYLSSLALLNSHFNYFKLKDVAFFILKCLSGKNPL 84
++P R DFQLQ E S ++L SHF Y+ D+A F+ L K PL
Sbjct 708 RVPHRIDFQLQPALTEK--SYWSVLKSHFAYWTNADLALFLANVLYCK-PL 755
> At1g31480
Length=869
Score = 34.7 bits (78), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 4/42 (9%)
Query 38 RYDFQLQEDTAEH-YLSSLALLNSHFNYFKLKDVAFFILKCL 78
R D LQE T EH YL ++ +H NY++ +D A FI+K L
Sbjct 764 RIDHMLQEKTFEHPYLQAIG---AHTNYWRDQDTALFIIKHL 802
> CE29636
Length=1119
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 38/82 (46%), Gaps = 11/82 (13%)
Query 5 TSSGLPIACGETQRAVSQLEAINAVKEEQQMPRRYDFQLQEDTAEHYLSSLALLNSHFNY 64
TS LPI + L + V + ++PRRY F+L D+ L + ++S +
Sbjct 703 TSLSLPIGFEDVD-----LYQVIVVHRDGRIPRRYGFRLSRDSKVGNLREVVAVSSGIS- 756
Query 65 FKLKDVAFFILKCLSGKNPLMS 86
++ ++C+S K LMS
Sbjct 757 -----MSHLTIQCMSSKGTLMS 773
> CE29635
Length=1116
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 38/82 (46%), Gaps = 11/82 (13%)
Query 5 TSSGLPIACGETQRAVSQLEAINAVKEEQQMPRRYDFQLQEDTAEHYLSSLALLNSHFNY 64
TS LPI + L + V + ++PRRY F+L D+ L + ++S +
Sbjct 700 TSLSLPIGFEDVD-----LYQVIVVHRDGRIPRRYGFRLSRDSKVGNLREVVAVSSGIS- 753
Query 65 FKLKDVAFFILKCLSGKNPLMS 86
++ ++C+S K LMS
Sbjct 754 -----MSHLTIQCMSSKGTLMS 770
> SPAC20G8.02
Length=757
Score = 33.5 bits (75), Expect = 0.16, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 38 RYDFQLQEDTAEHYLSSLALLNSHFNYFKLKDVAFFILKCL 78
R DF LQE + S ++ +N+H Y+K D+A FIL L
Sbjct 716 RIDFMLQEGALDTSYSYVSAMNAHSEYWKNVDLAHFILTQL 756
> Hs6005824
Length=1000
Score = 32.0 bits (71), Expect = 0.41, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 37 RRYDFQLQEDTAEHYLSSLALLNSHFNYFKLKDVAFFILK 76
RR D+ LQE E + L L SH Y++ +D A +LK
Sbjct 946 RRIDYVLQEKPIESFNEYLFALQSHLCYWESEDTALLLLK 985
> At2g34730
Length=829
Score = 30.8 bits (68), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 3 EETSSGLPIACGETQRAVSQLEAINAVK-EEQQMPRRYDFQLQEDT 47
EE+S+GL ET R +S E IN K E +M R +D+++QE T
Sbjct 308 EESSTGLIPEHNETLRHMSPDEMINHFKIEMNKMKRDHDYKIQELT 353
> CE15405
Length=219
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 6/60 (10%)
Query 78 LSGKNPL-----MSYTDHLQLLEAAARKAAEHAAKEGRPEEQQRQLQEAAALAERLKAFE 132
+GKNP+ SY D Q + R A +A +G+PEE+ ++++E + + K +E
Sbjct 86 FAGKNPIEEAQADSYIDQCQEYNTSFR-ACMYATLQGKPEEEVQKIREEVYIPAQNKFYE 144
> Hs20546650
Length=112
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 9/56 (16%)
Query 16 TQRAVSQLE-AINAVKEEQQMPRRYDFQLQEDTAEHYLSSLALLNSHFNYFKLKDV 70
TQ+ + Q+E IN + P+R D + +A+ Y L LLN HF YF K+V
Sbjct 15 TQKQMEQIEKMINNI------PKRQDRVSLDFSADSY--KLILLNGHFYYFIKKEV 62
> 7295225
Length=385
Score = 28.1 bits (61), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 28/64 (43%), Gaps = 5/64 (7%)
Query 11 IACGET---QRAVSQLEAINAVKEEQQMPRRYDFQLQEDTAEHYLSSLALLNSHFNYFKL 67
I CG V+QL V+ + + + D L D YL LAL SH N L
Sbjct 4 ILCGIVVLVTIVVNQLSEAREVRRRRGLMLQLDNYLSYDGIMQYLDELAL--SHSNRVTL 61
Query 68 KDVA 71
KDVA
Sbjct 62 KDVA 65
> SPAC328.06
Length=1141
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query 78 LSGKNPLMSYTDHLQLLE--AAARKAAEHAAKEGRPEEQQRQLQEAAALAERLKAFEASL 135
L+ + LMS D L +L AA ++ + P+ Q ++ A + LK EASL
Sbjct 985 LTTNSNLMSSVDTLSILSKWAAQQQDSRLPINPKLPDILQEEINNVVAEVDMLKKQEASL 1044
Query 136 EQPRKH 141
++ R H
Sbjct 1045 KEERTH 1050
> CE04481
Length=403
Score = 27.3 bits (59), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 34/73 (46%), Gaps = 9/73 (12%)
Query 57 LLNSHFNYFKLKDVAFFILKCLSGKNPLMSYTDHLQLLEAAARKAAEHAAKEGRPEEQQR 116
++N H +LK +L+ L G HL + E A R+ AE G P E++R
Sbjct 264 MINKHVECERLKPAKAEVLEKLKGH--------HLIMCELAYRRVAEIVKNVGGPTEKKR 315
Query 117 QLQEAAALAERLK 129
+E + ER++
Sbjct 316 -FEELSKTIERVQ 327
Lambda K H
0.314 0.126 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1712413322
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40