bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_3234_orf1
Length=86
Score E
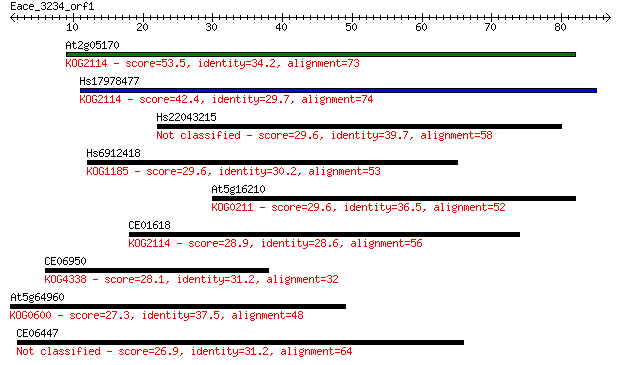
Sequences producing significant alignments: (Bits) Value
At2g05170 53.5 8e-08
Hs17978477 42.4 2e-04
Hs22043215 29.6 1.3
Hs6912418 29.6 1.5
At5g16210 29.6 1.5
CE01618 28.9 2.3
CE06950 28.1 3.7
At5g64960 27.3 6.5
CE06447 26.9 9.3
> At2g05170
Length=932
Score = 53.5 bits (127), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 47/73 (64%), Gaps = 0/73 (0%)
Query 9 RQIREVLMHIERHRLLPPLTVLEVLQQSPAVTLDAVKDYFLRASDELSEHLEQSQALLQS 68
++++EVL +IER +LPP+ VL+ L ++P +TL +KDY R ++ S+ +E+ + ++
Sbjct 756 KEVKEVLTYIERDDILPPIIVLQTLAKNPCLTLSVIKDYIARKLEQESKIIEEDRRAVEK 815
Query 69 DRAEVSYMAKEID 81
+ M KEI+
Sbjct 816 YQETTKNMRKEIE 828
> Hs17978477
Length=941
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 40/74 (54%), Gaps = 0/74 (0%)
Query 11 IREVLMHIERHRLLPPLTVLEVLQQSPAVTLDAVKDYFLRASDELSEHLEQSQALLQSDR 70
+ VL HIE L+PPL V++ L + TL ++DY ++ + S+ + Q + ++ R
Sbjct 738 VAAVLKHIENKNLMPPLLVVQTLAHNSTATLSVIRDYLVQKLQKQSQQIAQDELRVRRYR 797
Query 71 AEVSYMAKEIDRYK 84
E + + +EI K
Sbjct 798 EETTRIRQEIQELK 811
> Hs22043215
Length=268
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 32/58 (55%), Gaps = 7/58 (12%)
Query 22 RLLPPLTVLEVLQQSPAVTLDAVKDYFLRASDELSEHLEQSQALLQSDRAEVSYMAKE 79
RL P L +L PA+T + V D+ LRAS LS + +AL S RA +++A E
Sbjct 87 RLPPSLRLL------PAMTEEVVGDHHLRASKGLSNLNRRVRALAPS-RAGKAFVATE 137
> Hs6912418
Length=578
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 12 REVLMHIERHRLLPPLTVLEVLQQSPAVTLDAVKDYFLRASDELSEHLEQSQA 64
+E+L + ++PP+ +L +T K YF++ +EL + LEQS A
Sbjct 495 KEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLEQSLA 547
> At5g16210
Length=1189
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 31/55 (56%), Gaps = 3/55 (5%)
Query 30 LEVLQQSPAVTLDAVKDYFLRASDELSEHLEQSQALLQSD---RAEVSYMAKEID 81
L+V Q SPA DA++ Y+ + SE E+ A+LQ + + E+ ++KE D
Sbjct 183 LDVWQDSPAHVPDALRYYYYQYLSSTSEAAEEKIAMLQENESLKKEIERLSKEKD 237
> CE01618
Length=950
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 30/56 (53%), Gaps = 7/56 (12%)
Query 18 IERHRLLPPLTVLEVLQQSPAVTLDAVKDYFLRASDELSEHLEQSQALLQSDRAEV 73
IE + PL VLE+L ++ +T+ +V+DY + L + Q +++ DR +
Sbjct 696 IEASEQIHPLVVLELLAKNEHLTISSVRDYII-------AWLRKQQIIIEEDRNTI 744
> CE06950
Length=1613
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 10/32 (31%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 6 GMERQIREVLMHIERHRLLPPLTVLEVLQQSP 37
G + I+ +L+H+E +LP +L+ +Q++P
Sbjct 399 GTKNTIQHILVHMENEDILPLGQILKTIQETP 430
> At5g64960
Length=513
Score = 27.3 bits (59), Expect = 6.5, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 30/51 (58%), Gaps = 5/51 (9%)
Query 1 LASQKGMERQIREVLMHIERH--RLLPPLTVLEVLQQSPAV-TLDAVKDYF 48
+ S + ++R++RE+ H +RH LL + VL+ Q+ A LDA +YF
Sbjct 277 MKSSRPLKRRVREIYRHFDRHALELLEKMLVLDPSQRICAKDALDA--EYF 325
> CE06447
Length=652
Score = 26.9 bits (58), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 10/64 (15%)
Query 2 ASQKGMERQIREVLMHIERHRLLPPLTVLEVLQQSPAVTLDAVKDYFLRASDELSEHLEQ 61
A + G+++ +++VL H+E LLP +Q P V AV++ F+ ++E L++
Sbjct 208 AGKDGVQQCVQKVLDHLESKGLLP--------EQIPDVP--AVRELFVSDDLTVAELLKE 257
Query 62 SQAL 65
SQ L
Sbjct 258 SQNL 261
Lambda K H
0.318 0.131 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1190857334
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40